2025-12-04 京都大学
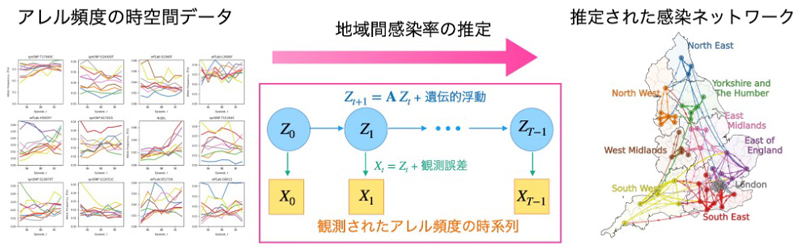
ウイルスのゲノムデータから地域間感染動態を推定する。
<関連情報>
- https://www.kyoto-u.ac.jp/ja/research-news/2025-12-04-0
- https://www.kyoto-u.ac.jp/sites/default/files/2025-12/web_2512_Okada-15f17d6ab0075da618c657fd067094af.pdf
- https://www.pnas.org/doi/10.1073/pnas.2500663122
中立アレル頻度の時系列から異質なコミュニティ間疾患伝播を明らかにする Uncovering heterogeneous intercommunity disease transmission from neutral allele frequency time series
Takashi Okada, Giulio Isacchini, QinQin Yu, and Oskar Hallatschek
Proceedings of the National Academy of Sciences Published:November 26, 2025
DOI:https://doi.org/10.1073/pnas.2500663122
Significance
Current methods for understanding disease transmission between communities rely on indirect data, such as mobility patterns and contact surveys, which introduce significant uncertainty-especially when regions differ in policies, immunity, and travel patterns. To overcome these limitations, we introduce a computational framework that directly infers disease importation rates using genomic data alone. Applied to SARS-CoV-2, our method reveals transmission pathways, highlighting the importance of long-range interactions and their shifts across variant waves. By revealing how infections move among communities, this method enhances epidemic forecasting and explains why certain regions contribute disproportionately to pathogen evolution. While further validation is needed, it may also be applicable beyond SARS-CoV-2, including to other genomic data such as microbiome or ancient DNA studies.
Abstract
The COVID-19 pandemic has underscored the need for accurate epidemic forecasting to predict pathogen spread, evolution, and evaluate intervention strategies. Forecast reliability hinges on detailed knowledge of disease transmission across population segments, which may be inferred from contact surveys or mobility data. However, these indirect approaches make it difficult to estimate rare transmissions between socially or geographically distant communities. We show that the steep ramp-up of genome sequencing surveillance during the pandemic can be leveraged to directly identify transmission patterns between geographically defined communities. Our approach uses a hidden Markov model to infer the fraction of infections a community imports from others based on how rapidly allele frequencies in the focal community converge to those in the donor communities. Applying this method to SARS-CoV-2 sequencing data from England and the United States, we uncover networks of intercommunity transmission that reflect geographical relationships while exposing significant long-range interactions. The scaling of importation rate with distance is consistent across both countries, yet weaker than expected based on mobility data, highlighting limitations of indirect inference. We show that transmission patterns can change between waves of variants of concern and analyze how the inferred heterogeneity in intercommunity transmission impacts evolutionary forecasts. While applied here to geographically defined communities, our approach could be applied to those defined by other traits (e.g., age, socioeconomic status), provided time-series data can be stratified accordingly. Overall, our study highlights population genomic time series data as a crucial record of epidemiological interactions, which can be deciphered using tree-free inference methods.