2026-01-06 マサチューセッツ工科大学(MIT)
<関連情報>
- https://news.mit.edu/2026/ai-generated-sensors-open-new-paths-early-cancer-detection-0106
- https://www.nature.com/articles/s41467-025-67226-1
- https://www.nature.com/articles/nbt.2464
ディープラーニングによるプロテアーゼ基質の設計 Deep learning guided design of protease substrates
Carmen Martin-Alonso,Sarah Alamdari,Tahoura S. Samad,Kevin K. Yang,Sangeeta N. Bhatia & Ava P. Amini
Nature Communications Published:06 January 2026
DOI:https://doi.org/10.1038/s41467-025-67226-1
Abstract
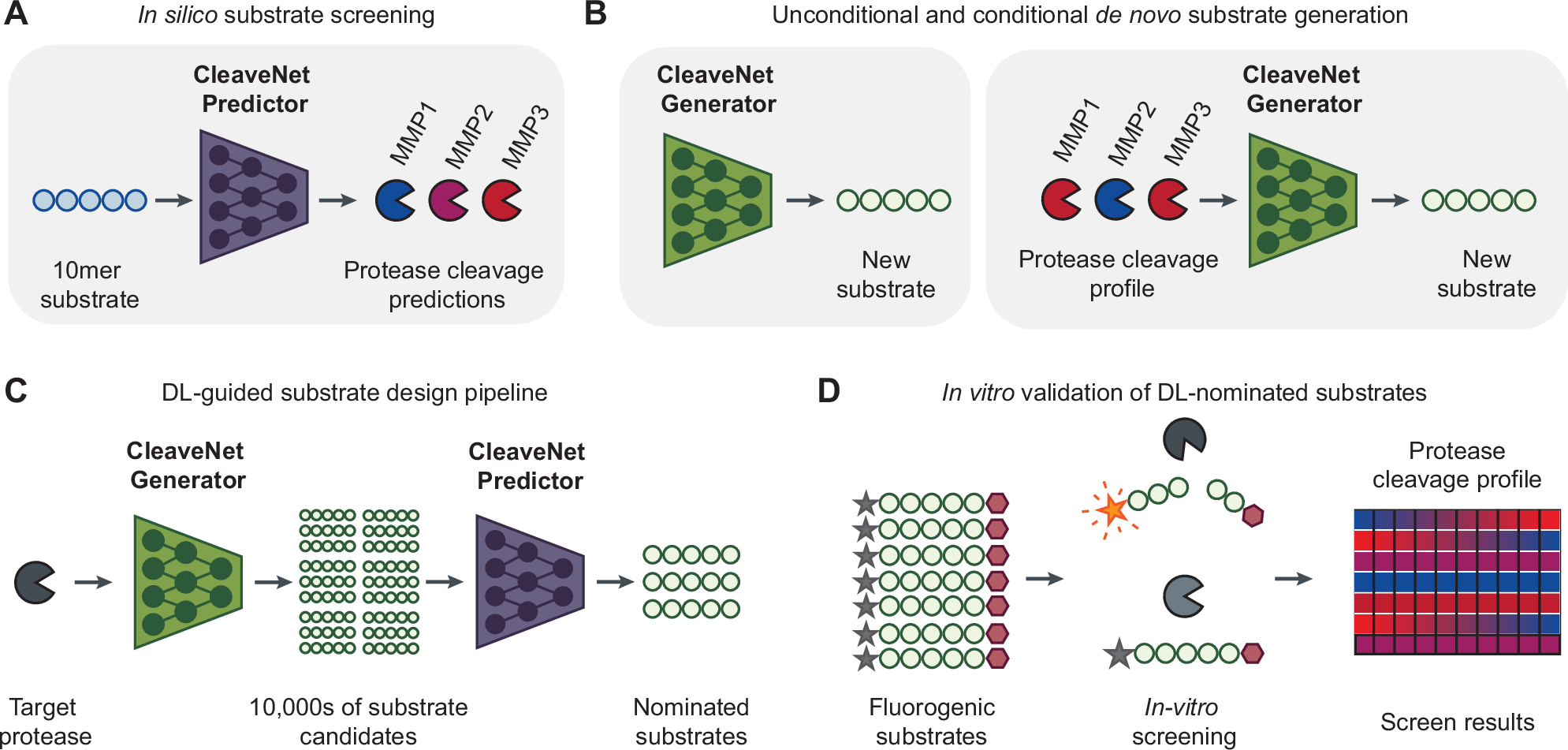
Proteases, enzymes that play critical roles in health and disease, exert their function through the cleavage of peptide bonds. Identifying substrates that are efficiently and selectively cleaved by target proteases is essential for studying protease activity and for harnessing it in protease-activated diagnostics and therapeutics. However, the vast design space of possible substrates (c.a. 2010 amino acid combinations for a 10-mer peptide) and the limited accessibility of high-throughput activity profiling tools hinder the speed and success of substrate design. We present CleaveNet, an end-to-end AI pipeline for the design of protease substrates. Applied to matrix metalloproteinases, CleaveNet enhances the scale, tunability, and efficiency of substrate design. CleaveNet generates peptide substrates that exhibit sound biophysical properties and capture not only well-established but also previously-uncharacterized cleavage motifs. To control substrate design, CleaveNet incorporates a conditioning tag that steers peptide generation towards desired cleavage profiles, enabling targeted design of efficient and selective substrates. CleaveNet-generated substrates were validated experimentally through a large-scale in vitro screen, even in the challenging case of designing highly selective substrates for MMP13. We envision that CleaveNet will accelerate our ability to study and capitalize on protease activity, paving the way for in silico design tools across enzyme classes.
多重尿疾患モニタリングのための質量コード化合成バイオマーカー Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease
Gabriel A Kwong,Geoffrey von Maltzahn,Gayathree Murugappan,Omar Abudayyeh,Steven Mo,Ioannis A Papayannopoulos,Deanna Y Sverdlov,Susan B Liu,Andrew D Warren,Yury Popov,Detlef Schuppan & Sangeeta N Bhatia
Nature Biotechnology Published:16 December 2012
DOI:https://doi.org/10.1038/nbt.2464
Abstract
Biomarkers are becoming increasingly important in the clinical management of complex diseases, yet our ability to discover new biomarkers remains limited by our dependence on endogenous molecules. Here we describe the development of exogenously administered ‘synthetic biomarkers’ composed of mass-encoded peptides conjugated to nanoparticles that leverage intrinsic features of human disease and physiology for noninvasive urinary monitoring. These protease-sensitive agents perform three functions in vivo: they target sites of disease, sample dysregulated protease activities and emit mass-encoded reporters into host urine for multiplexed detection by mass spectrometry. Using mouse models of liver fibrosis and cancer, we show that these agents can noninvasively monitor liver fibrosis and resolution without the need for invasive core biopsies and substantially improve early detection of cancer compared with current clinically used blood biomarkers. This approach of engineering synthetic biomarkers for multiplexed urinary monitoring should be broadly amenable to additional pathophysiological processes and point-of-care diagnostics.