2026-01-15 ワシントン大学セントルイス校
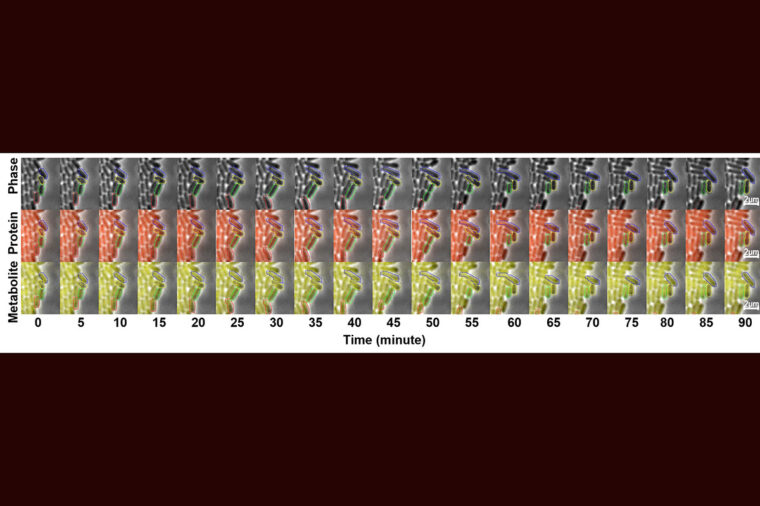
WashU researchers track single cells to reveal enzyme copy number fluctuation as the main source of metabolic noise. (Photo: Alex Schmitz and Xinyue Mu)
<関連情報>
- https://source.washu.edu/2026/01/exploring-metabolic-noise-opens-new-paths-to-better-biomanufacturing/
- https://www.nature.com/articles/s41467-025-67733-1
大腸菌におけるベタキサンチン生産増強のための単一細胞生合成ノイズとダイナミクスの探究 Exploring single-cell biosynthetic noise and dynamics for enhanced betaxanthin production in Escherichia coli
Xinyue Mu,Alexander C. Schmitz,Zhenya Ding,Wei Li,Abhyudai Singh & Fuzhong Zhang
Nature Communications Published:21 December 2025
DOI:https://doi.org/10.1038/s41467-025-67733-1 An unedited version of this manuscript
Abstract
Cell-to-cell variability often limits the efficiency of microbial bioproduction, yet how individual cells fluctuate over time and how these fluctuations shape population-level output remain unclear. To address this issue, we tracked a heterologous betaxanthin pathway in Escherichia coli using microfluidics-assisted time-lapse microscopy, allowing simultaneous measurement of fluctuations in betaxanthin, its biosynthetic enzyme DOD and growth across generations. Here we show that over 50% of high betaxanthin producers become medium or low producers after two divisions. Betaxanthin variation primarily originates from DOD noise, with a smaller contribution from growth rate fluctuations. We further develop a stochastic model to explore various control circuits and find that pathway enzyme or metabolite-based growth selection strategies are most effective in enhancing production. We experimentally validate the model by coupling enzyme expression to nutrient availability, which enriches high producers and boosts titer by 4.4-fold. Our results highlight key sources of metabolic heterogeneity and provide a framework for designing robust microbial processes.