2026-02-04 フランス国立科学研究センター(CNRS)
<関連情報>
- https://www.cnrs.fr/en/press/3d-architecture-genome-enables-cells-remember-their-past
- https://www.nature.com/articles/s41588-025-02489-4
一過性のヒストン脱アセチル化酵素阻害は遺伝子発現と3Dゲノムフォールディングの細胞記憶を誘導する Transient histone deacetylase inhibition induces cellular memory of gene expression and 3D genome folding
Flora Paldi,Michael-Florian Szalay,Solène Dufau,Marco Di Stefano,Hadrien Reboul,Daniel Jost,Frédéric Bantignies & Giacomo Cavalli
Nature Genetics Published:04 February 2026
DOI:https://doi.org/10.1038/s41588-025-02489-4
Abstract
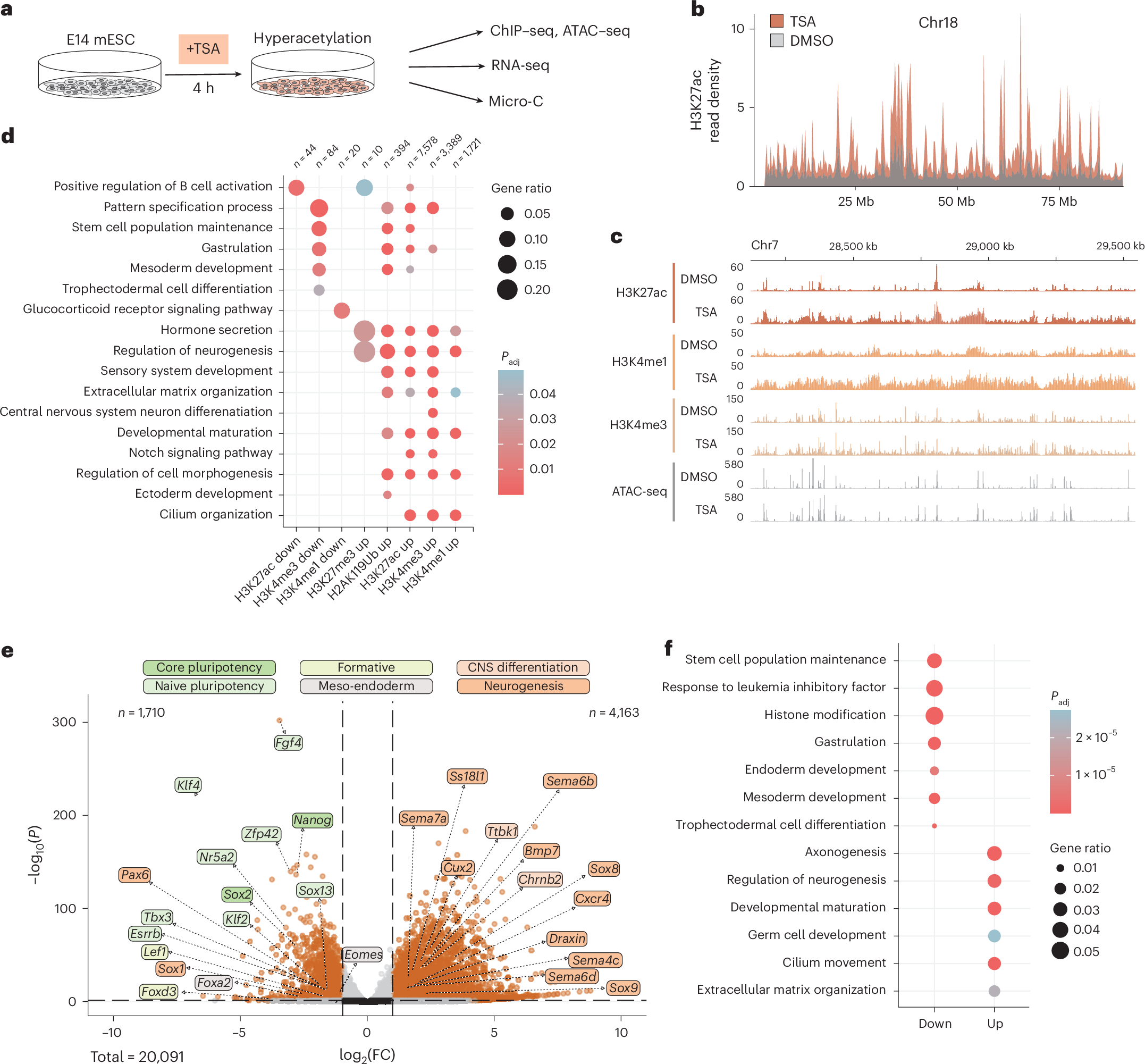
Epigenetic memory enables the propagation of gene expression patterns following transient stimuli. Although three-dimensional chromatin organization is emerging as a key regulator of genome function, it is unknown whether it contributes to cellular memory. Here we establish that acute perturbation of the epigenome can induce cellular memory of gene expression in mouse embryonic stem cells. We uncover how a pulse of histone deacetylase inhibition translates to changes in transcription, histone modifications and genome folding. While most epigenomic and transcriptional changes are initially reversed once the perturbation is removed, some loci remain transcriptionally deregulated and genome architecture partially maintains its perturbed conformation. Consequently, a second pulse of transient hyperacetylation induces stronger memory of transcriptional deregulation. Using ultradeep Micro-C, we associate memory of gene expression with repressive Polycomb-mediated chromatin topology. These results demonstrate how cells can record transient stresses in their genome architecture, thereby enabling an enhanced response to subsequent perturbations.