適応策や回復力のある作物の開発には、このメカニズムが重要である可能性があります。 Mechanism could be important for adaptation, development of resilient crops
2022-06-13 ペンシルベニア州立大学(PennState)
研究チームは、シロイヌナズナにおけるリボスニッチの存在を実験的に証明した後、配列が決定された数百種類のシロイヌナズナのゲノムからリボスニッチの可能性を予測する大規模な計算機研究を実施した。その結果、評価対象となった380万以上のSNPのうち、100万以上、つまり約27%がリボスニッチとして機能する可能性があることがわかった。
<関連情報>
- https://www.psu.edu/news/eberly-college-science/story/climate-associated-genetic-switches-found-plants/
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02656-4
シロイヌナズナにおける気候関連リボスニッチの実験的実証と全構造体予測 Experimental demonstration and pan-structurome prediction of climate-associated riboSNitches in Arabidopsis
Ángel Ferrero-Serrano,Megan M. Sylvia,Peter C. Forstmeier,Andrew J. Olson,Doreen Ware,Philip C. Bevilacqua & Sarah M. Assmann
Genome Biology Published:19 April 2022
DOI:https://doi.org/10.1186/s13059-022-02656-4
Abstract
Background
Genome-wide association studies (GWAS) aim to correlate phenotypic changes with genotypic variation. Upon transcription, single nucleotide variants (SNVs) may alter mRNA structure, with potential impacts on transcript stability, macromolecular interactions, and translation. However, plant genomes have not been assessed for the presence of these structure-altering polymorphisms or “riboSNitches.”
Results
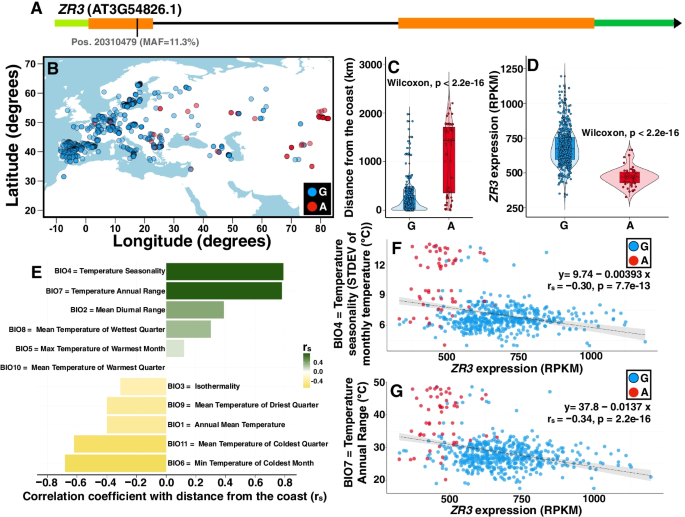
We experimentally demonstrate the presence of riboSNitches in transcripts of two Arabidopsis genes, ZINC RIBBON 3 (ZR3) and COTTON GOLGI-RELATED 3 (CGR3), which are associated with continentality and temperature variation in the natural environment. These riboSNitches are also associated with differences in the abundance of their respective transcripts, implying a role in regulating the gene’s expression in adaptation to local climate conditions. We then computationally predict riboSNitches transcriptome-wide in mRNAs of 879 naturally inbred Arabidopsis accessions. We characterize correlations between SNPs/riboSNitches in these accessions and 434 climate descriptors of their local environments, suggesting a role of these variants in local adaptation. We integrate this information in CLIMtools V2.0 and provide a new web resource, T-CLIM, that reveals associations between transcript abundance variation and local environmental variation.
Conclusion
We functionally validate two plant riboSNitches and, for the first time, demonstrate riboSNitch conditionality dependent on temperature, coining the term “conditional riboSNitch.” We provide the first pan-genome-wide prediction of riboSNitches in plants. We expand our previous CLIMtools web resource with riboSNitch information and with 1868 additional Arabidopsis genomes and 269 additional climate conditions, which will greatly facilitate in silico studies of natural genetic variation, its phenotypic consequences, and its role in local adaptation.


