光を取り込むタンパク質に照らされた数十億年前の地球 Earth of billions of years ago illuminated by light-capturing proteins
2022-06-27 カリフォルニア大学リバーサイド校(UCR)
ロドプシンは、太陽光をエネルギーに変換する能力をもつタンパク質で、そのエネルギーを使って細胞内のプロセスを活性化させることができます。
ロドプシンは、人間の目の杆体や錐体と関係があり、明暗を区別したり色を見たりすることができる。また、現代の生物や、鮮やかな虹色を呈する塩田などの環境にも広く分布している。
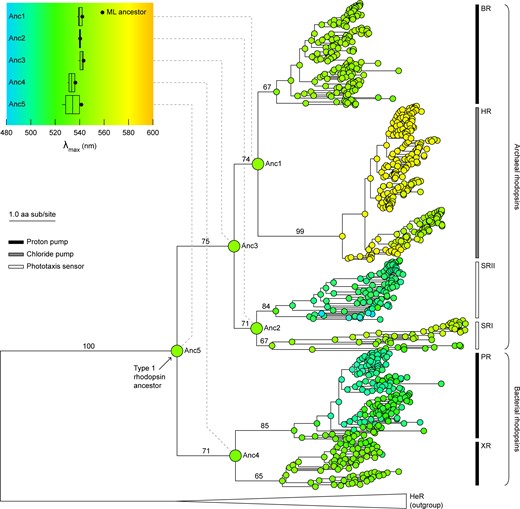
研究チームは、機械学習を用いて世界中のロドプシンタンパク質の配列を解析し、それらが時間とともにどのように進化してきたかを追跡した。そして、一種の家系図を作成し、25億年から40億年前のロドプシンや、ロドプシンが直面していたと思われる条件を復元することに成功したのです。
<関連情報>
- https://news.ucr.edu/articles/2022/06/27/ancient-microbes-may-help-us-find-extraterrestrial-life-forms
- https://academic.oup.com/mbe/article/39/5/msac100/6582242?searchresult=1&login=false#356974379
太古の微生物ロドプシンが示す最古の光帯ニッチ Earliest Photic Zone Niches Probed by Ancestral Microbial Rhodopsins
Cathryn D. Sephus, Evrim Fer, Amanda K. Garcia, Zachary R. Adam, Edward W. Schwieterman, Betul Kacar
Molecular Biologyand Evolution Published::07 May 2022
DOI:https://doi.org/10.1093/molbev/msac100
Abstract
For billions of years, life has continuously adapted to dynamic physical conditions near the Earth’s surface. Fossils and other preserved biosignatures in the paleontological record are the most direct evidence for reconstructing the broad historical contours of this adaptive interplay. However, biosignatures dating to Earth’s earliest history are exceedingly rare. Here, we combine phylogenetic inference of primordial rhodopsin proteins with modeled spectral features of the Precambrian Earth environment to reconstruct the paleobiological history of this essential family of photoactive transmembrane proteins. Our results suggest that ancestral microbial rhodopsins likely acted as light-driven proton pumps and were spectrally tuned toward the absorption of green light, which would have enabled their hosts to occupy depths in a water column or biofilm where UV wavelengths were attenuated. Subsequent diversification of rhodopsin functions and peak absorption frequencies was enabled by the expansion of surface ecological niches induced by the accumulation of atmospheric oxygen. Inferred ancestors retain distinct associations between extant functions and peak absorption frequencies. Our findings suggest that novel information encoded by biomolecules can be used as “paleosensors” for conditions of ancient, inhabited niches of host organisms not represented elsewhere in the paleontological record. The coupling of functional diversification and spectral tuning of this taxonomically diverse protein family underscores the utility of rhodopsins as universal testbeds for inferring remotely detectable biosignatures on inhabited planetary bodies.