2万個の遺伝子配列解析による2段階アプローチにより、免疫チェックポイント阻害剤の投与対象者の予測精度が向上 A two-step approach with sequencing of 20,000 genes improves prediction of who benefits from immune checkpoint inhibitors
2022-07-08 ニューヨーク大学 (NYU)
全エクソーム解析は、ゲノムの2パーセントに当たる約2万個のタンパク質をコードする遺伝子の配列を決定し、疾患に関与している可能性のある変異を探す方法です。
全エクソームシーケンスはがん治療にはあまり使用されていませんが、最近、免疫療法に関するいくつかの研究でシーケンスが使用され始めています。これらの研究は小規模ではあるが、ゲノム因子と免疫療法に対する患者の反応との関係を明らかにするのに役立つと考えられる。
<関連情報>
- https://www.nyu.edu/about/news-publications/news/2022/july/whole-exome-sequencing-cancer-immunotherapy.html
- https://www.nature.com/articles/s41467-022-31055-3
免疫療法効果の予測因子としての再発性体細胞変異 Recurrent somatic mutations as predictors of immunotherapy response
Zoran Z. Gajic,Aditya Deshpande,Mateusz Legut,Marcin Imieliński &Neville E. Sanjana
Nature Communications Published:08 July 2022
DOI:https://doi.org/10.1038/s41467-022-31055-3
Abstract
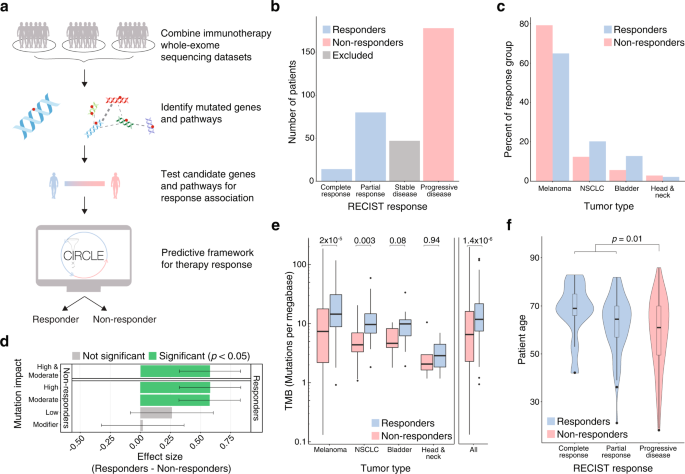
Immune checkpoint blockade (ICB) has transformed the treatment of metastatic cancer but is hindered by variable response rates. A key unmet need is the identification of biomarkers that predict treatment response. To address this, we analyzed six whole exome sequencing cohorts with matched disease outcomes to identify genes and pathways predictive of ICB response. To increase detection power, we focus on genes and pathways that are significantly mutated following correction for epigenetic, replication timing, and sequence-based covariates. Using this technique, we identify several genes (BCLAF1, KRAS, BRAF, and TP53) and pathways (MAPK signaling, p53 associated, and immunomodulatory) as predictors of ICB response and develop the Cancer Immunotherapy Response CLassifiEr (CIRCLE). Compared to tumor mutational burden alone, CIRCLE led to superior prediction of ICB response with a 10.5% increase in sensitivity and a 11% increase in specificity. We envision that CIRCLE and more broadly the analysis of recurrently mutated cancer genes will pave the way for better prognostic tools for cancer immunotherapy.