2023-07-07 オークリッジ国立研究所(ORNL)
◆この研究により、TFIIHは形状を変えることができることがわかりました。この研究は、遺伝性疾患の原因となる変異による遺伝子疾患の起源を理解するために、転写とDNA修復のインターフェースにおけるTFIIHの内部機構を解明することが重要です。この研究は、論文「Nature Communications」に掲載されました。
<関連情報>
- https://www.ornl.gov/news/georgia-state-researchers-use-ornl-supercomputer-gain-new-insights-dna-repair
- https://www.nature.com/articles/s41467-023-38416-6
転写とDNA修復におけるTFIIHの機能の根底にあるダイナミックなコンフォメーションスイッチングが遺伝病に影響を与える Dynamic conformational switching underlies TFIIH function in transcription and DNA repair and impacts genetic diseases
Jina Yu,Chunli Yan,Thomas Dodd,Chi-Lin Tsai,John A. Tainer,Susan E. Tsutakawa & Ivaylo Ivanov
Nature Communications Published:13 May 2023
DOI:https://doi.org/10.1038/s41467-023-38416-6
Abstract
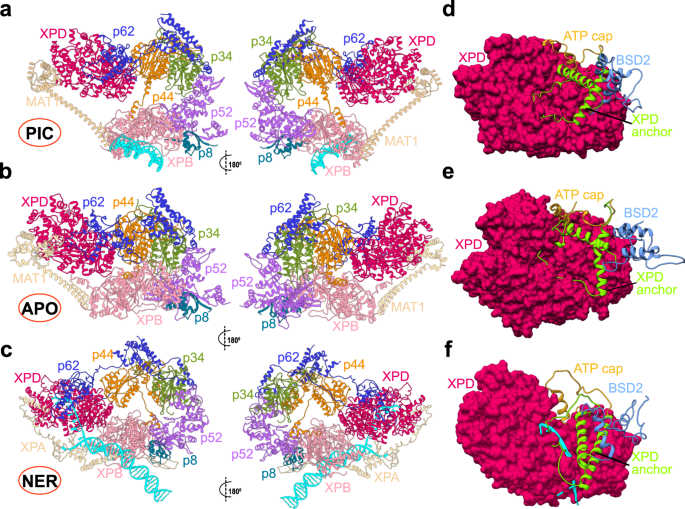
Transcription factor IIH (TFIIH) is a protein assembly essential for transcription initiation and nucleotide excision repair (NER). Yet, understanding of the conformational switching underpinning these diverse TFIIH functions remains fragmentary. TFIIH mechanisms critically depend on two translocase subunits, XPB and XPD. To unravel their functions and regulation, we build cryo-EM based TFIIH models in transcription- and NER-competent states. Using simulations and graph-theoretical analysis methods, we reveal TFIIH’s global motions, define TFIIH partitioning into dynamic communities and show how TFIIH reshapes itself and self-regulates depending on functional context. Our study uncovers an internal regulatory mechanism that switches XPB and XPD activities making them mutually exclusive between NER and transcription initiation. By sequentially coordinating the XPB and XPD DNA-unwinding activities, the switch ensures precise DNA incision in NER. Mapping TFIIH disease mutations onto network models reveals clustering into distinct mechanistic classes, affecting translocase functions, protein interactions and interface dynamics.