2023-12-14 カリフォルニア大学サンディエゴ校(UCSD)
◆研究はBrain Initiativeの一環で、脳の基本状態を把握し、神経・精神疾患に対する新しい治療法を模索することを目指しています。これにより、脳の特定の部位が特定の疾患にどのように影響されるかを理解し、より精確でターゲット指向の治療法の開発に寄与する可能性があります。
<関連情報>
- https://today.ucsd.edu/story/mapping-the-mouse-brain-helps-reveal-what-makes-us-human
- https://www.nature.com/articles/s41586-023-06824-9
- https://www.nature.com/articles/s41586-023-06819-6
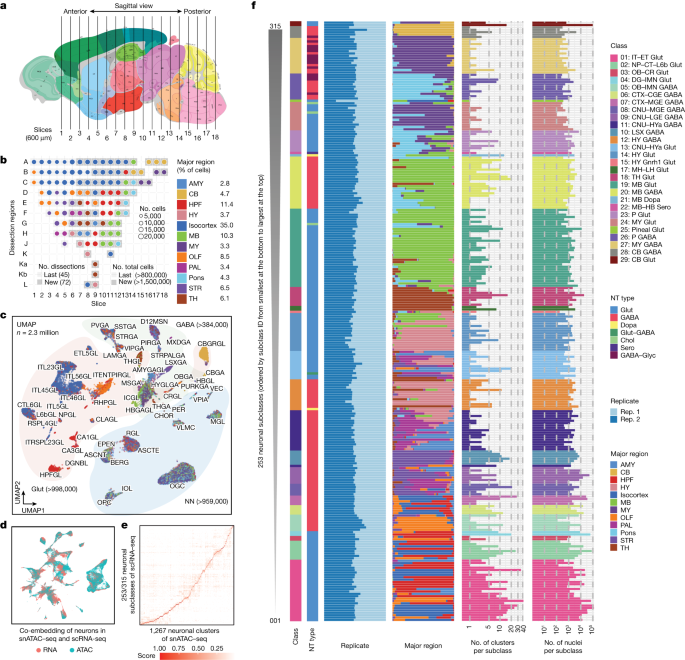
成体マウスの脳におけるクロマチンへのアクセス性の単一細胞分析 Single-cell analysis of chromatin accessibility in the adult mouse brain
Songpeng Zu,Yang Eric Li,Kangli Wang,Ethan J. Armand,Sainath Mamde,Maria Luisa Amaral,Yuelai Wang,Andre Chu,Yang Xie,Michael Miller,Jie Xu,Zhaoning Wang,Kai Zhang,Bojing Jia,Xiaomeng Hou,Lin Lin,Qian Yang,Seoyeon Lee,Bin Li,Samantha Kuan,Hanqing Liu,Jingtian Zhou,Antonio Pinto-Duarte,Jacinta Lucero,Julia Osteen,Michael Nunn,Kimberly A. Smith,Bosiljka Tasic,Zizhen Yao,Hongkui Zeng,Zihan Wang,Jingbo Shang,M. Margarita Behrens,Joseph R. Ecker,Allen Wang,Sebastian Preissl & Bing Ren
Nature Published:13 December 2023
DOI:https://doi.org/10.1038/s41586-023-06824-9
Abstract
Recent advances in single-cell technologies have led to the discovery of thousands of brain cell types; however, our understanding of the gene regulatory programs in these cell types is far from complete1,2,3,4. Here we report a comprehensive atlas of candidate cis-regulatory DNA elements (cCREs) in the adult mouse brain, generated by analysing chromatin accessibility in 2.3 million individual brain cells from 117 anatomical dissections. The atlas includes approximately 1 million cCREs and their chromatin accessibility across 1,482 distinct brain cell populations, adding over 446,000 cCREs to the most recent such annotation in the mouse genome. The mouse brain cCREs are moderately conserved in the human brain. The mouse-specific cCREs—specifically, those identified from a subset of cortical excitatory neurons—are strongly enriched for transposable elements, suggesting a potential role for transposable elements in the emergence of new regulatory programs and neuronal diversity. Finally, we infer the gene regulatory networks in over 260 subclasses of mouse brain cells and develop deep-learning models to predict the activities of gene regulatory elements in different brain cell types from the DNA sequence alone. Our results provide a resource for the analysis of cell-type-specific gene regulation programs in both mouse and human brains.
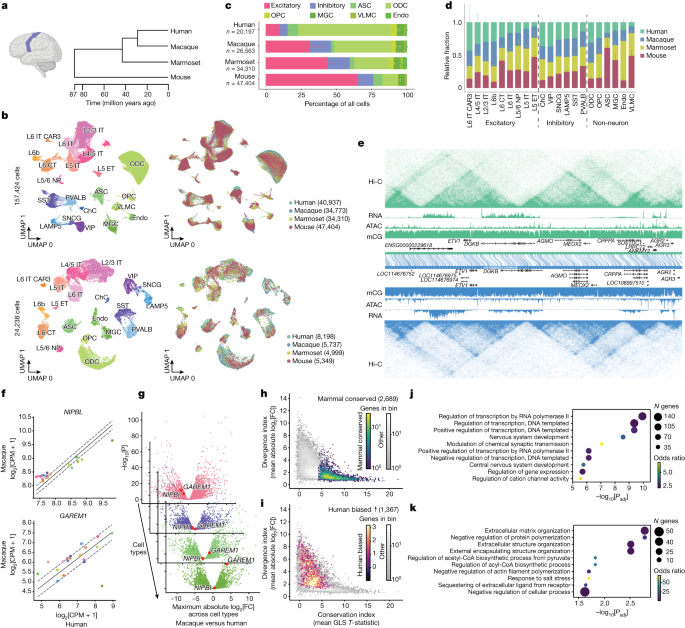
哺乳類新皮質の保存された多様な遺伝子制御プログラム Conserved and divergent gene regulatory programs of the mammalian neocortex
Nathan R. Zemke,Ethan J. Armand,Wenliang Wang,Seoyeon Lee,Jingtian Zhou,Yang Eric Li,Hanqing Liu,Wei Tian,Joseph R. Nery,Rosa G. Castanon,Anna Bartlett,Julia K. Osteen,Daofeng Li,Xiaoyu Zhuo,Vincent Xu,Lei Chang,Keyi Dong,Hannah S. Indralingam,Jonathan A. Rink,Yang Xie,Michael Miller,Fenna M. Krienen,Qiangge Zhang,Naz Taskin,Jonathan Ting,Guoping Feng,Steven A. McCarroll,Edward M. Callaway,Ting Wang,Ed S. Lein,M. Margarita Behrens,Joseph R. Ecker & Bing Ren
Nature Published:13 December 2023
DOI:https://doi.org/10.1038/s41586-023-06819-6
Abstract
Divergence of cis-regulatory elements drives species-specific traits1, but how this manifests in the evolution of the neocortex at the molecular and cellular level remains unclear. Here we investigated the gene regulatory programs in the primary motor cortex of human, macaque, marmoset and mouse using single-cell multiomics assays, generating gene expression, chromatin accessibility, DNA methylome and chromosomal conformation profiles from a total of over 200,000 cells. From these data, we show evidence that divergence of transcription factor expression corresponds to species-specific epigenome landscapes. We find that conserved and divergent gene regulatory features are reflected in the evolution of the three-dimensional genome. Transposable elements contribute to nearly 80% of the human-specific candidate cis-regulatory elements in cortical cells. Through machine learning, we develop sequence-based predictors of candidate cis-regulatory elements in different species and demonstrate that the genomic regulatory syntax is highly preserved from rodents to primates. Finally, we show that epigenetic conservation combined with sequence similarity helps to uncover functional cis-regulatory elements and enhances our ability to interpret genetic variants contributing to neurological disease and traits.