2024-07-31 テキサス大学オースチン校(UT Austin)
<関連情報>
- https://news.utexas.edu/2024/07/31/ai-opens-door-to-safe-effective-new-antibiotics-to-combat-resistant-bacteria/
- https://www.nature.com/articles/s41551-024-01243-1
- https://www.cell.com/cell/fulltext/S0092-8674(17)31451-4
膜選択性を駆動する抗菌ペプチドの特徴解析のための Deep mutational scanning and machine learning for the analysis of antimicrobial-peptide features driving membrane selectivity
Justin R. Randall,Luiz C. Vieira,Claus O. Wilke & Bryan W. Davies
Nature Biomedical Engineering Published:31 July 2024
DOI:https://doi.org/10.1038/s41551-024-01243-1
Abstract
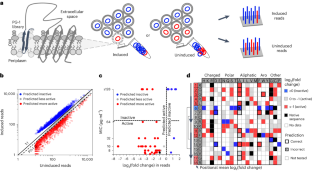
Many antimicrobial peptides directly disrupt bacterial membranes yet can also damage mammalian membranes. It is therefore central to their therapeutic use that rules governing the membrane selectivity of antimicrobial peptides be deciphered. However, this is difficult even for short peptides owing to the large combinatorial space of amino acid sequences. Here we describe a method for measuring the loss or maintenance of antimicrobial-peptide activity for thousands of peptide-sequence variants simultaneously, and its application to Protegrin-1, a potent yet toxic antimicrobial peptide, to determine the positional importance and flexibility of residues across its sequence while identifying variants with changes in membrane selectivity. More bacterially selective variants maintained a membrane-bound secondary structure while avoiding aromatic residues and cysteine pairs. A machine-learning model trained with our datasets accurately predicted membrane-specific activities for over 5.7 million Protegrin-1 variants, and identified one variant that showed substantially reduced toxicity and retention of activity in a mouse model of intraperitoneal infection. The high-throughput methodology may help elucidate sequence–structure–function relationships in antimicrobial peptides and inform the design of peptide-based synthetic drugs.
表面表示ペプチドライブラリーの細菌セルフスクリーニングによる次世代抗菌薬の発見 Discovery of Next-Generation Antimicrobials through Bacterial Self-Screening of Surface-Displayed Peptide Libraries
Ashley T. Tucker,Sean P. Leonard,Cory D. DuBois,…,Claus O. Wilke,M. Stephen Trent,Bryan W. Davies
Cell Published:January 04, 2018
DOI:https://doi.org/10.1016/j.cell.2017.12.009
Graphical Abstract

Video Abstract
(mp4, (17.45 MB)
Highlights
- Development of a high-throughput platform for discovery of antimicrobial peptides
- Screening 800,000 peptides uncovered thousands of synthetic antimicrobial sequences
- Lead peptides exhibit potent antimicrobial activity and distinctive mechanisms
- Lead hit antimicrobial physicochemistry extends far beyond what nature has evolved
Summary
Peptides have great potential to combat antibiotic resistance. While many platforms can screen peptides for their ability to bind to target cells, there are virtually no platforms that directly assess the functionality of peptides. This limitation is exacerbated when identifying antimicrobial peptides because the phenotype, death, selects against itself and has caused a scientific bottleneck that confines research to a few naturally occurring classes of antimicrobial peptides. We have used this seeming dissonance to develop Surface Localized Antimicrobial Display (SLAY), a platform that allows screening of unlimited numbers of peptides of any length, composition, and structure in a single tube for antimicrobial activity. Using SLAY, we screened ∼800,000 random peptide sequences for antimicrobial function and identified thousands of active sequences, dramatically increasing the number of known antimicrobial sequences. SLAY hits present with different potential mechanisms of peptide action and access to areas of antimicrobial physicochemical space beyond what nature has evolved.