2024-08-08 スイス連邦工科大学ローザンヌ校(EPFL)
<関連情報>
- https://actu.epfl.ch/news/scientists-unlock-the-secrets-to-an-alpine-flower-/
- https://onlinelibrary.wiley.com/doi/10.1111/eva.13737
- https://www.mdpi.com/2072-4292/13/8/1588
遺伝子型-環境相関研究における超高解像度環境プロキシの統合 Integrating very high resolution environmental proxies in genotype–environment association studies
Annie S. Guillaume, Kevin Leempoel, Aude Rogivue, Felix Gugerli, Christian Parisod, Stéphane Joost
Evolutionary Applications Published: 28 June 2024
DOI:https://doi.org/10.1111/eva.13737

Abstract
Landscape genomic analyses associating genetic variation with environmental variables are powerful tools for studying molecular signatures of species’ local adaptation and for detecting candidate genes under selection. The development of landscape genomics over the past decade has been spurred by improvements in resolutions of genomic and environmental datasets, allegedly increasing the power to identify putative genes underlying local adaptation in non-model organisms. Although these associations have been successfully applied to numerous species across a diverse array of taxa, the spatial scale of environmental predictor variables has been largely overlooked, potentially limiting conclusions to be reached with these methods. To address this knowledge gap, we systematically evaluated performances of genotype–environment association (GEA) models using predictor variables at multiple spatial resolutions. Specifically, we used multivariate redundancy analyses to associate whole-genome sequence data from the plant Arabis alpina L. collected across four neighboring valleys in the western Swiss Alps, with very high-resolution topographic variables derived from digital elevation models of grain sizes between 0.5 m and 16 m. These comparisons highlight the sensitivity of landscape genomic models to spatial resolution, where the optimal grain sizes were specific to variable type, terrain characteristics, and study extent. To assist in selecting variables at appropriate spatial resolutions, we demonstrate a practical approach to produce, select, and integrate multiscale variables into GEA models. After generalizing fine-grained variables to multiple spatial resolutions, a forward selection procedure is applied to retain only the most relevant variables for a particular context. Depending on the spatial resolution, the relevance for topographic variables in GEA studies calls for integrating multiple spatial scales into landscape genomic models. By carefully considering spatial resolutions, candidate genes under selection by a more realistic range of pressures can be detected for downstream analyses, with important applied implications for experimental research and conservation management of natural populations.
高山生態学におけるマルチスケール超高解像度地形モデル: 空中LiDARとドローンベースのステレオ写真測量技術の長所と短所 Multiscale Very High Resolution Topographic Models in Alpine Ecology: Pros and Cons of Airborne LiDAR and Drone-Based Stereo-Photogrammetry Technologies
Annie S. Guillaume,Kevin Leempoel,,Estelle Rochat,Aude Rogivue,Michel Kasser,Felix Gugerli,Christian Parisod andStéphane Joost
Remote Sensing Published: 20 April 2021
DOI:https://doi.org/10.3390/rs13081588
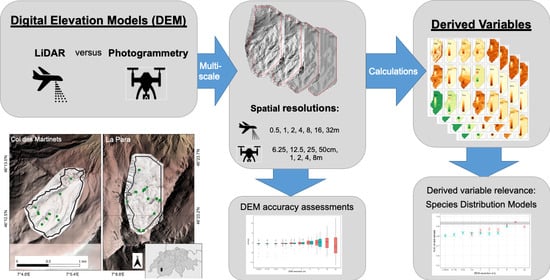
Graphical Abstract
Abstract
The vulnerability of alpine environments to climate change presses an urgent need to accurately model and understand these ecosystems. Popularity in the use of digital elevation models (DEMs) to derive proxy environmental variables has increased over the past decade, particularly as DEMs are relatively cheaply acquired at very high resolutions (VHR; <1 m spatial resolution). Here, we implement a multiscale framework and compare DEM-derived variables produced by Light Detection and Ranging (LiDAR) and stereo-photogrammetry (PHOTO) methods, with the aim of assessing their relevance and utility in species distribution modelling (SDM). Using a case study on the arctic-alpine plant, Arabis alpina, in two valleys in the western Swiss Alps, we show that both LiDAR and PHOTO technologies can be relevant for producing DEM-derived variables for use in SDMs. We demonstrate that PHOTO DEMs, up to a spatial resolution of at least 1 m, rivalled the accuracy of LiDAR DEMs, largely owing to the customizability of PHOTO DEMs to the study sites compared to commercially available LiDAR DEMs. We obtained DEMs at spatial resolutions of 6.25 cm–8 m for PHOTO and 50 cm–32 m for LiDAR, where we determined that the optimal spatial resolutions of DEM-derived variables in SDM were between 1 and 32 m, depending on the variable and site characteristics. We found that the reduced extent of PHOTO DEMs altered the calculations of all derived variables, which had particular consequences on their relevance at the site with heterogenous terrain. However, for the homogenous site, SDMs based on PHOTO-derived variables generally had higher predictive powers than those derived from LiDAR at matching resolutions. From our results, we recommend carefully considering the required DEM extent to produce relevant derived variables. We also advocate implementing a multiscale framework to appropriately assess the ecological relevance of derived variables, where we caution against the use of VHR-DEMs finer than 50 cm in such studies.