2024-10-22 ニューヨーク大学 (NYU)
<関連情報>
- https://www.nyu.edu/about/news-publications/news/2024/october/stirred–not-shaken-scientists-uncover-how-transcription-drives-.html
- https://www.nature.com/articles/s41467-024-51149-4
- https://www.pnas.org/doi/full/10.1073/pnas.1220313110
ヒト生細胞における単一遺伝子の転写依存的移動とゲノム全体の運動 Transcription-dependent mobility of single genes and genome-wide motions in live human cells
Fang-Yi Chu,Alexis S. Clavijo,Suho Lee & Alexandra Zidovska
Nature Communications Published:22 October 2024
DOI:https://doi.org/10.1038/s41467-024-51149-4
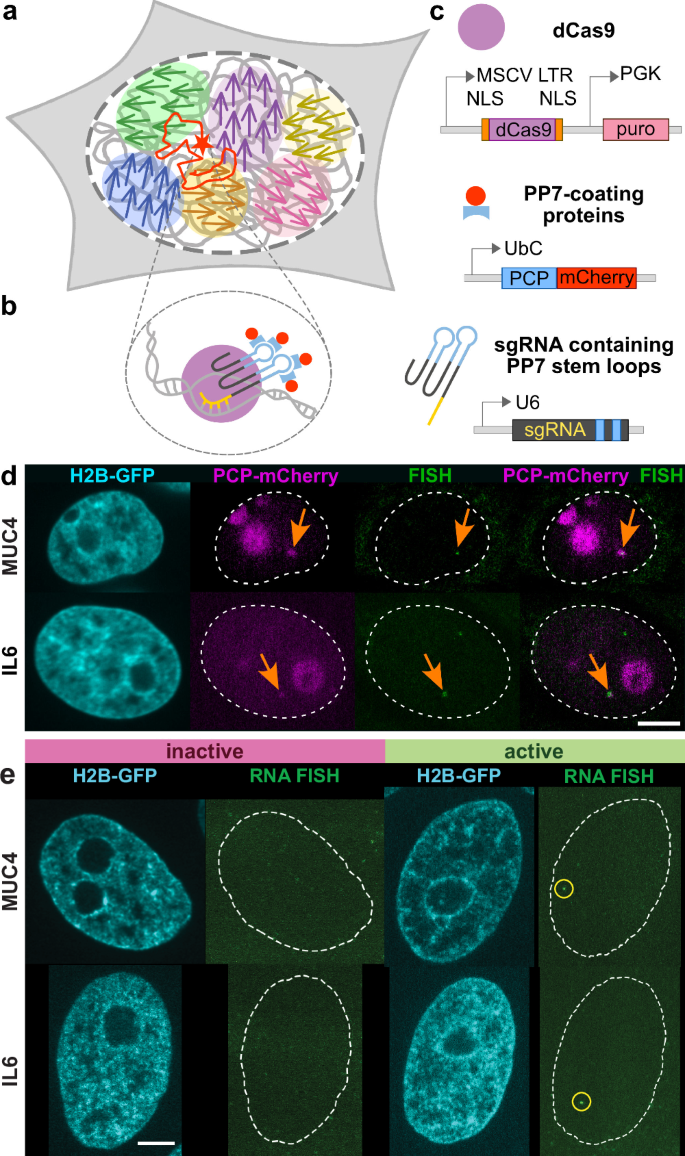
Abstract
The human genome is highly dynamic across all scales. At the gene level, chromatin is persistently remodeled and rearranged during active processes such as transcription, replication and DNA repair. At the genome level, chromatin moves in micron-scale domains that break up and re-form over seconds, but the origin of these coherent motions is unknown. Here, we investigate the connection between genomic motions and gene-level activity. Simultaneous mapping of single-gene and genome-wide motions shows that the coupling of gene transcriptional activity to flows of the nearby genome is modulated by chromatin compaction. A motion correlation analysis suggests that a single active gene drives larger-scale motions in low-compaction regions, but high-compaction chromatin drives gene motion regardless of its activity state. By revealing unexpected connections among gene activity, spatial heterogeneities of chromatin and its emergent genome-wide motions, these findings uncover aspects of the genome’s spatiotemporal organization that directly impact gene regulation and expression.
間期クロマチンダイナミクスにおけるミクロンスケールのコヒーレンス Micron-scale coherence in interphase chromatin dynamics
Alexandra Zidovska, David A. Weitz, and Timothy J. Mitchison
Proceedings of the National Academy of Sciences Published:September 9, 2013
DOI:https://doi.org/10.1073/pnas.1220313110

Significance
Chromatin, the functional form of DNA inside the cell nucleus, has been heavily studied using biochemical and genetic methods, but we still know little about its large-scale organization and even less about its dynamics. We present a unique method, displacement correlation spectroscopy (DCS), which allows us to map interphase chromatin dynamics simultaneously across the entire nucleus and follow the temporal evolution of the global chromatin dynamics in vivo. Using DCS we discovered that chromatin moves coherently across micron-scale regions for a few seconds, which implies a transient mechanical coupling between chromatin on different chromosomes. This coupling might allow different regions of the nucleus to communicate by a unique, mechanochemical mechanism, e.g., to coordinate responses to DNA damage.
Abstract
Chromatin structure and dynamics control all aspects of DNA biology yet are poorly understood, especially at large length scales. We developed an approach, displacement correlation spectroscopy based on time-resolved image correlation analysis, to map chromatin dynamics simultaneously across the whole nucleus in cultured human cells. This method revealed that chromatin movement was coherent across large regions (4–5 µm) for several seconds. Regions of coherent motion extended beyond the boundaries of single-chromosome territories, suggesting elastic coupling of motion over length scales much larger than those of genes. These large-scale, coupled motions were ATP dependent and unidirectional for several seconds, perhaps accounting for ATP-dependent directed movement of single genes. Perturbation of major nuclear ATPases such as DNA polymerase, RNA polymerase II, and topoisomerase II eliminated micron-scale coherence, while causing rapid, local movement to increase; i.e., local motions accelerated but became uncoupled from their neighbors. We observe similar trends in chromatin dynamics upon inducing a direct DNA damage; thus we hypothesize that this may be due to DNA damage responses that physically relax chromatin and block long-distance communication of forces.


