2025-03-13 マウントサイナイ医療システム (MSHS)
<関連情報>
- https://www.mountsinai.org/about/newsroom/2025/mount-sinai-researchers-develop-method-to-identify-dormant-cells-that-carry-hiv
- https://www.nature.com/articles/s41467-025-57368-7
リザーバー標識ヒト化マウスモデルを用いたART治療初期のHIV-1感染におけるT細胞系譜を越えたHIVの持続性の追跡 Tracking HIV persistence across T cell lineages during early ART-treated HIV-1-infection using a reservoir-marking humanized mouse model
Namita Satija,Foramben Patel,Gerrit Schmidt,Donald V. Doanman,Manav Kapoor,Annalena La Porte,Ying-Chih Wang,Kenneth M. Law,Anthony M. Esposito,Kimaada Allette,Kristin G. Beaumont,Robert P. Sebra & Benjamin K. Chen
Nature Communications Published:06 March 2025
DOI:https://doi.org/10.1038/s41467-025-57368-7
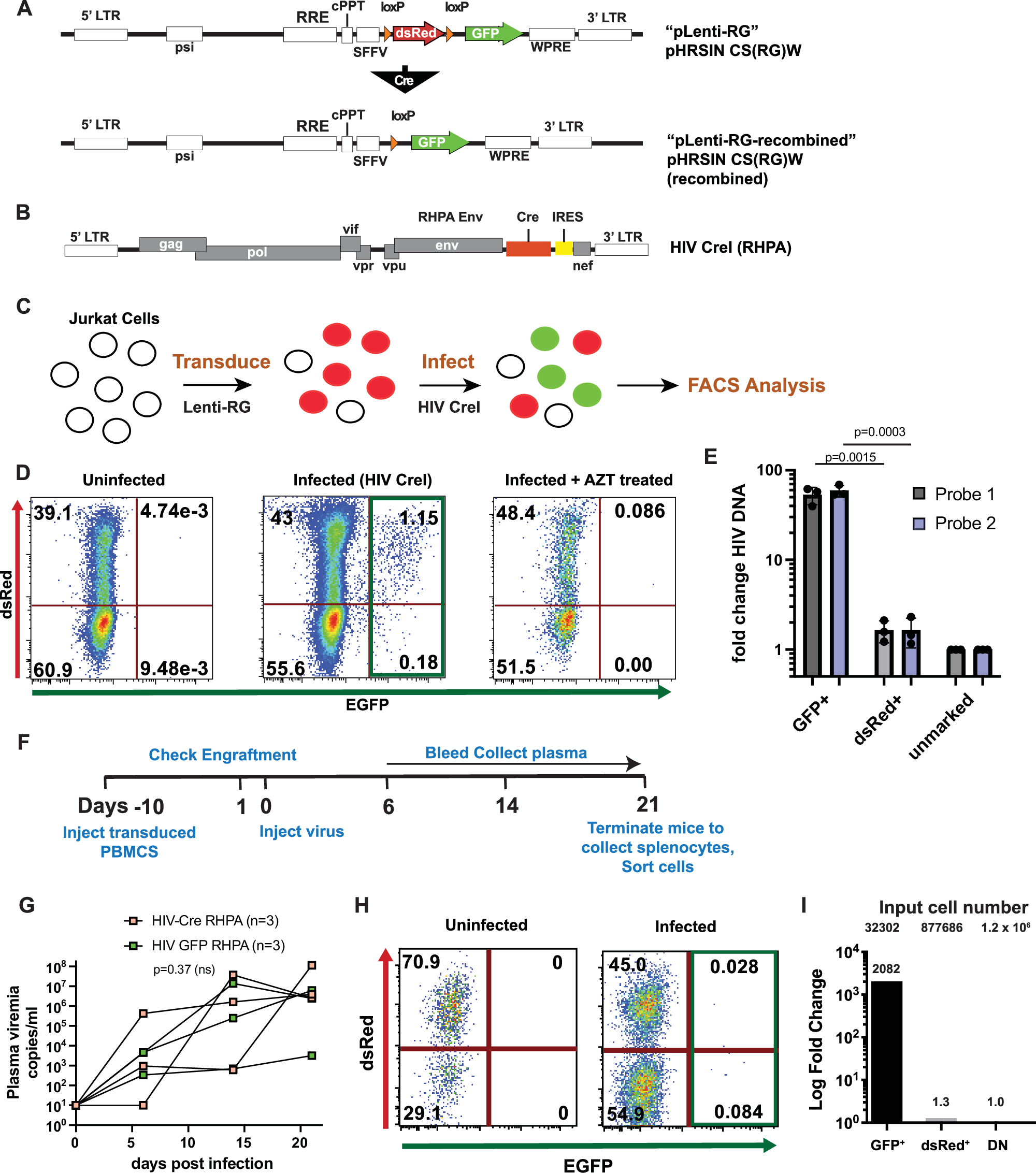
Abstract
Human immunodeficiency virus (HIV) infection depletes CD4 T-cells, and long-term persistence of latent virus prevents full clearance of HIV even in the presence of effective antiretroviral therapy (ART), Here we present the HIV-1-induced lineage tracing (HILT) system, a model that irreversibly marks infected cells within a humanized mouse model, which detects rare latently infected cells. Immunodeficient mice transplanted with genetically modified hematopoietic stem cells develop a human immune system, in which CD4 T-cells contain a genetic switch that permanently labels cells infected by HIV-1 expressing cre-recombinase. Through single-cell RNA sequencing of HILT-marked cells during acute infection and post-ART treatment, we identify distinct CD4+ T-cell transcriptional lineages enriched in either active or latent infections. Comparative gene expression analysis highlights common pathways modulated in both states, including EIF2, Sirtuin, and protein ubiquitination. Critical regulators of these pathways, including JUN, BCL2, and MDM2, change to opposite directions in the two states, highlighting gene expression programs that may support HIV persistence across T-cell lineages and states.