2025-08-25 国立遺伝学研究所
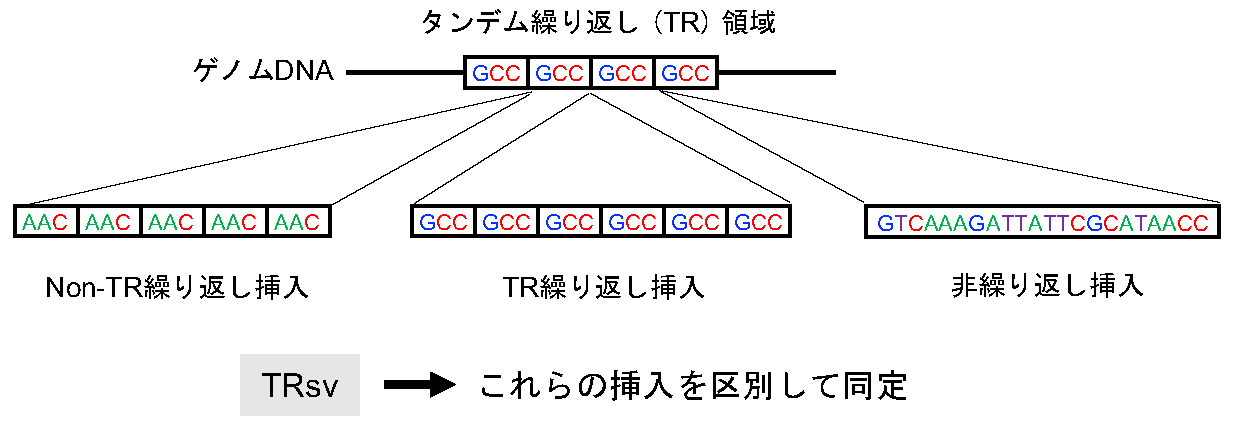
図1: TRsvはタンデム繰り返し領域で観察される異なるタイプの挿入を検出する
タンデム繰り返し(TR)領域内では、TR領域の繰り返し単位(図の例ではGCC)と同じ繰り返し単位からなるTR繰り返し挿入が観察されることが多いが、異なる繰り返し単位からなる挿入(Non-TR繰り返し挿入)や、繰り返しを持たない挿入(非繰り返し挿入)がしばしば観察される。TRsvは、これらの異なるタイプの挿入を区別して同定する。
<関連情報>
- https://www.nig.ac.jp/nig/ja/2025/08/research-highlights_ja/pr20250825.html
- https://www.nig.ac.jp/nig/images/research_highlights/PR20250820.pdf
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03718-z
TRsv:ロングリードシーケンスデータを用いたタンデムリピート変異、構造変異、および短いインデルの同時検出 TRsv: simultaneous detection of tandem repeat variations, structural variations, and short indels using long read sequencing data
Shunichi Kosugi & Chikashi Terao
Genome Biology Published:20 August 2025
DOI:https://doi.org/10.1186/s13059-025-03718-z
Abstract
Tandem repeat copy number variations (TR-CNVs), structural variations (SVs), and short indels have been responsible for many diseases and traits, but no tools exist to distinguish and detect these variants. In this study, we developed a computational tool, TRsv, to distinguish and detect TR-CNVs, SVs, and short indels using long reads. In evaluation with simulated and real datasets, TRsv outperformed existing tools for detection of TR-CNVs and indels and performed equally well for detection of SVs. We demonstrated genome-wide detection of TR-CNVs, including variants associated with gene expression, disease, and quantitative traits, using 160 long-read whole genome sequencing data and TRsv.

