2025-09-04 カリフォルニア大学バークレー校(UCB)
Web要約 の発言:
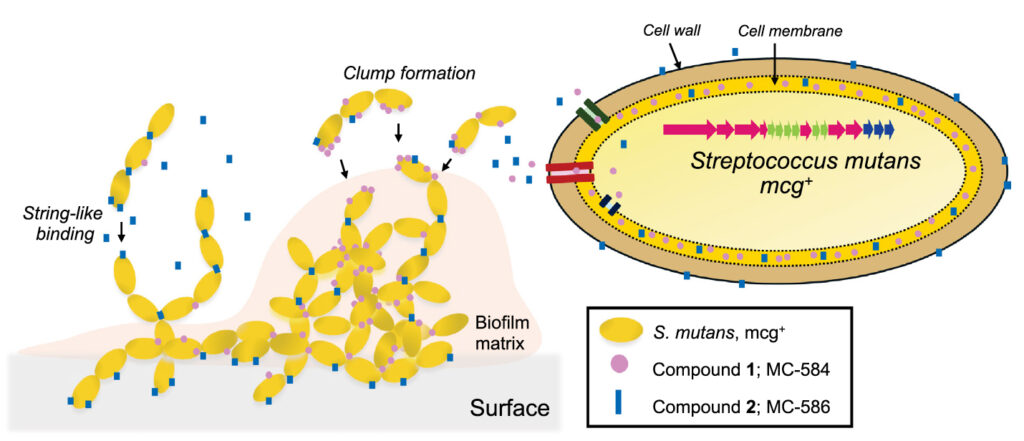 A graphic showing how two different small molecules (blue and pink) made by bacteria in the mouth help oral bacteria (yellow) form strands and clumps, respectively, in a biofilm that sticks to teeth. One known cavity-causing bacteria, Streptococcus mutans (right), contains a cluster of genes that makes these molecules. Mapping the many molecules synthesized by oral bacteria may pinpoint gene clusters that can help the good bacteria in the mouth out-compete the bad and prevent cavities.
A graphic showing how two different small molecules (blue and pink) made by bacteria in the mouth help oral bacteria (yellow) form strands and clumps, respectively, in a biofilm that sticks to teeth. One known cavity-causing bacteria, Streptococcus mutans (right), contains a cluster of genes that makes these molecules. Mapping the many molecules synthesized by oral bacteria may pinpoint gene clusters that can help the good bacteria in the mouth out-compete the bad and prevent cavities.
Wenjun Zhang and McKenna Yao/UC Berkeley
<関連情報>
- https://news.berkeley.edu/2025/09/04/can-the-good-bacteria-in-your-mouth-act-as-probiotic-cavity-fighters/
- https://www.pnas.org/doi/10.1073/pnas.2504492122
ヒト口腔内微生物叢における分岐した生合成経路由来の特異的代謝物の相乗作用 Synergistic action of specialized metabolites from divergent biosynthesis in the human oral microbiome
McKenna Loop Yao, Nicholas A. Zill, Colin Charles Barber, +5 , and Wenjun Zhang
Proceedings of the National Academy of Sciences August 19, 2025
DOI:https://doi.org/10.1073/pnas.2504492122
Significance
Dental cavities, or caries, are one of the most common health issues worldwide, yet the factors that drive harmful bacteria to slime our teeth are not fully understood. Our research reveals that the oral bacteria responsible for cavities produce specialized compounds that play a key role in forming decay-causing biofilms. Using computational microbiome analysis to correlate dental caries with specialized genes from oral bacterial samples, we identified a unique set of bacterial genes that create two molecules, called mutanoclumpins, which work together to promote biofilm formation. By understanding the synthesis and function of these molecules, we provide insights into the hidden chemistry of the oral microbiome, which could lead to innovative ways to prevent or treat cavities.
Abstract
Despite extensive efforts, our understanding of the virulence factors contributing to oral biofilm formation—a hallmark of dental caries—remains incomplete. We present evidence that the specialized metabolism of the oral microbiome is a critical yet underexplored factor in oral biofilm formation. Through microbiome analysis, we identified a hybrid nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) encoding biosynthetic gene cluster that correlates with dental caries and is widely represented in oral pathogens, including Streptococcus mutans. This gene cluster produces two major mutanoclumpin metabolites, MC-584 and MC-586, which feature molecular scaffolds differing in a C–C macrocyclic linkage. Both metabolites synergistically promote robust biofilm formation of S. mutans through a rare dual-metabolite mode of action. Further, each metabolite binds uniquely to the S. mutans cell surface, resulting in distinct multicellular morphologies. The biosynthesis of mutanoclumpins employs a unique chemical logic that produces two major products, rare within PKS-NRPS assembly lines. This study underscores the importance of characterizing genes implicated in human diseases through microbiome analysis and lays the foundation for exploring strategies to inhibit streptococci-induced dental caries.


