2026-01-29 東京大学,大阪大学,理化学研究所,愛知県がんセンター,国立がん研究センター
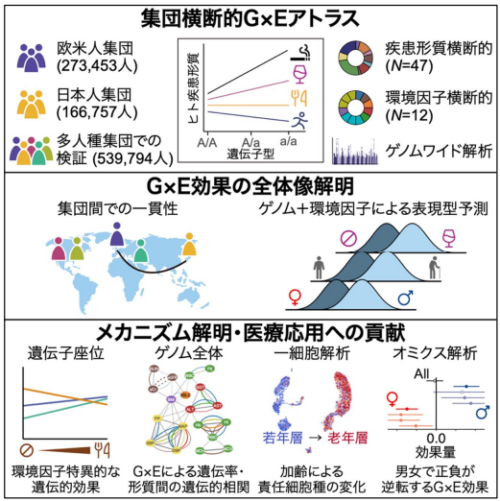
大規模解析により G×E 効果の全体像を解明
ワイン、食事、運動、性別のアイコンの出典:https://phosphoricons.com/; MIT license
<関連情報>
遺伝子と環境の相互作用に関する集団横断的な概要 A cross-population compendium of gene–environment interactions
Shinichi Namba,Kyuto Sonehara,Yuriko N. Koyanagi,Takezo Kikuchi,Takafumi Ojima,Ryuya Edahiro,Go Sato,Taiki Yamaji,Yoshihiko Tomofuji,Hiroyuki Ueda,Kenichi Yamamoto,Yosuke Ogawa,Ken Suzuki,Akinori Kanai,Shinichi Higashiue,Shuzo Kobayashi,Hiroki Yamaguchi,Yasunobu Nagata,Yasushi Okazaki,Naoyuki Matsumoto,Kenta Motomura,Hidenobu Koga,Asahi Hishida,Hiroaki Ikezaki,the BioBank Japan Project,… Yukinori Okada
Nature Published:28 January 2026
DOI:https://doi.org/10.1038/s41586-025-10054-6
Abstract
Environmental differences in genetic effect sizes, namely, gene–environment interactions, may uncover the genetic encoding of phenotypic plasticity1,2,3. We provide a cross-population atlas of gene–environment interactions comprising 440,210 individuals from European and Japanese populations, with replication in 539,794 individuals from diverse populations. By decomposing the contributions from age, sex and lifestyles, we delineate the aetiology of these gene–environment interactions, including a reverse-causality from a disease-related dietary change. Genome-wide analyses uncovered missing heritability and trait–trait relationships connected by the synergistic effects of genome and environments, which systematically affected polygenic prediction accuracy and cross-population portability. Single-cell projection revealed aging shift of pathways and cell types responsible for genetic regulation. Omics-level gene–environment analyses identified multiple sex-discordant genetic effects in lipid metabolism, informing clinical trial failures for genetically supported drug development. Our comprehensive gene–environment study decodes the dynamics of genetic associations, offering insights into complex trait biology, personalized medicine and drug development.