2023-06-13 カリフォルニア大学バークレー校(UCB)
◆この研究は、米国カリフォルニア大学バークレー校を中心とした2,000万ドルの研究プロジェクトであり、3つの新しい論文がその目標に向けた重要な進展を報告しています。最終目標は、翻訳システムを完全にプログラム可能にすることであり、新しい建築ブロック(現在のアミノ酸ではないもの)をリボソームに供給することによって、リボソームが無制限の新しい分子鎖の多様性を生み出せるようにすることです。
◆これらの分子鎖は、新しい生体材料、新しい酵素、さらには新しい薬物の基礎となる可能性があります。
<関連情報>
- https://news.berkeley.edu/2023/06/13/retooling-the-translation-machine-could-expand-the-chemical-repertoire-of-cells/
- https://www.nature.com/articles/s41557-023-01226-w
- https://pubs.acs.org/doi/10.1021/acscentsci.3c00153
- https://www.nature.com/articles/s41557-023-01224-y
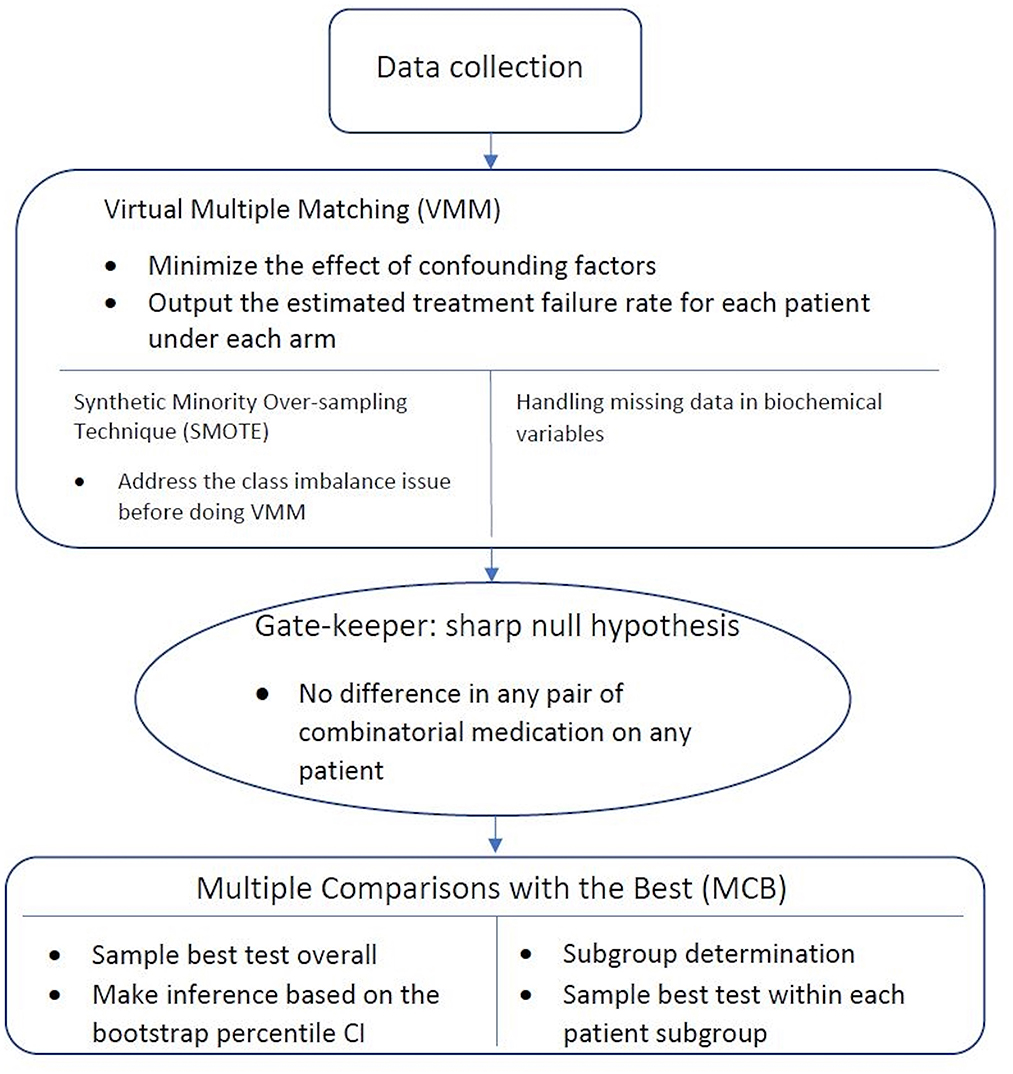
大腸菌リボソームの原子論的シミュレーションにより、翻訳活性基質の選択基準が示される Atomistic simulations of the Escherichia coli ribosome provide selection criteria for translationally active substrates
Zoe L. Watson,Isaac J. Knudson,Fred R. Ward,Scott J. Miller,Jamie H. D. Cate,Alanna Schepartz & Ara M. Abramyan
Nature Chemistry Published:12 June 2023
DOI:https://doi.org/10.1038/s41557-023-01226-w
Abstract
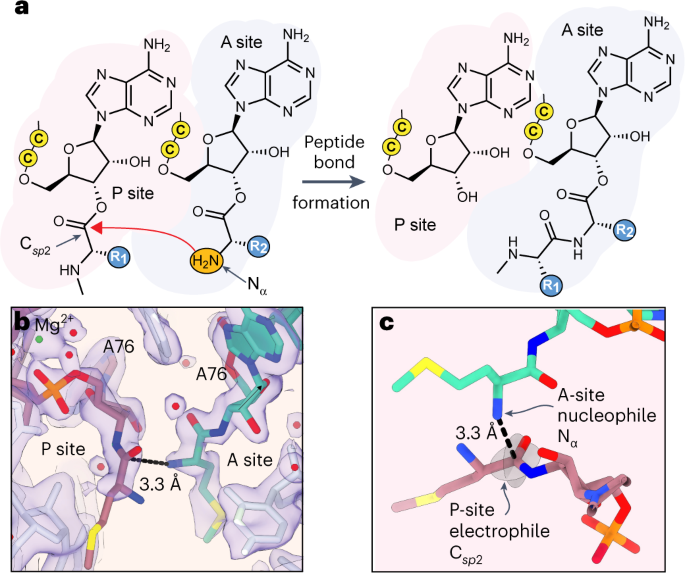
As genetic code expansion advances beyond L-α-amino acids to backbone modifications and new polymerization chemistries, delineating what substrates the ribosome can accommodate remains a challenge. The Escherichia coli ribosome tolerates non-L-α-amino acids in vitro, but few structural insights that explain how are available, and the boundary conditions for efficient bond formation are so far unknown. Here we determine a high-resolution cryogenic electron microscopy structure of the E. coli ribosome containing α-amino acid monomers and use metadynamics simulations to define energy surface minima and understand incorporation efficiencies. Reactive monomers across diverse structural classes favour a conformational space where the aminoacyl-tRNA nucleophile is <4 Å from the peptidyl-tRNA carbonyl with a Bürgi–Dunitz angle of 76–115°. Monomers with free energy minima that fall outside this conformational space do not react efficiently. This insight should accelerate the in vivo and in vitro ribosomal synthesis of sequence-defined, non-peptide heterooligomers.
アミノ安息香酸誘導体がリボソームの触媒センターにおける誘引フィットを阻害する Aminobenzoic Acid Derivatives Obstruct Induced Fit in the Catalytic Center of the Ribosome
Chandrima Majumdar, Joshua A. Walker, Matthew B. Francis, Alanna Schepartz, and Jamie H. D. Cate
ACS Central Science Published:May 30, 2023
DOI:https://doi.org/10.1021/acscentsci.3c00153

Abstract
The Escherichia coli (E. coli) ribosome can incorporate a variety of non-l-α-amino acid monomers into polypeptide chains in vitro but with poor efficiency. Although these monomers span a diverse set of compounds, there exists no high-resolution structural information regarding their positioning within the catalytic center of the ribosome, the peptidyl transferase center (PTC). Thus, details regarding the mechanism of amide bond formation and the structural basis for differences and defects in incorporation efficiency remain unknown. Within a set of three aminobenzoic acid derivatives─3-aminopyridine-4-carboxylic acid (Apy), ortho-aminobenzoic acid (oABZ), and meta-aminobenzoic acid (mABZ)─the ribosome incorporates Apy into polypeptide chains with the highest efficiency, followed by oABZ and then mABZ, a trend that does not track with the nucleophilicity of the reactive amines. Here, we report high-resolution cryo-EM structures of the ribosome with each of these three aminobenzoic acid derivatives charged on tRNA bound in the aminoacyl-tRNA site (A-site). The structures reveal how the aromatic ring of each monomer sterically blocks the positioning of nucleotide U2506, thereby preventing rearrangement of nucleotide U2585 and the resulting induced fit in the PTC required for efficient amide bond formation. They also reveal disruptions to the bound water network that is believed to facilitate formation and breakdown of the tetrahedral intermediate. Together, the cryo-EM structures reported here provide a mechanistic rationale for differences in reactivity of aminobenzoic acid derivatives relative to l-α-amino acids and each other and identify stereochemical constraints on the size and geometry of non-monomers that can be accepted efficiently by wild-type ribosomes.
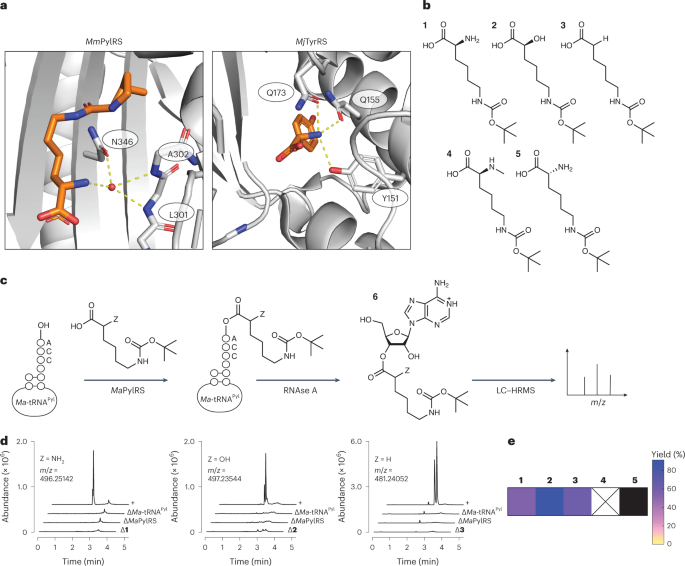
ピロリシル基転移型RNA合成酵素の基質範囲を拡大し、in vitroおよびin vivoで非α-アミノ酸を含むようになる Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo
Riley Fricke,Cameron V. Swenson,Leah Tang Roe,Noah Xue Hamlish,Bhavana Shah,Zhongqi Zhang,Elise Ficaretta,Omer Ad,Sarah Smaga,Christine L. Gee,Abhishek Chatterjee & Alanna Schepartz
Nature Chemistry Published:01 June 2023
DOI:https://doi.org/10.1038/s41557-023-01224-y
Abstract
The absence of orthogonal aminoacyl-transfer RNA (tRNA) synthetases that accept non-L-α-amino acids is a primary bottleneck hindering the in vivo translation of sequence-defined hetero-oligomers and biomaterials. Here we report that pyrrolysyl-tRNA synthetase (PylRS) and certain PylRS variants accept α-hydroxy, α-thio and N-formyl-L-α-amino acids, as well as α-carboxy acid monomers that are precursors to polyketide natural products. These monomers are accommodated and accepted by the translation apparatus in vitro; those with reactive nucleophiles are incorporated into proteins in vivo. High-resolution structural analysis of the complex formed between one PylRS enzyme and a m-substituted 2-benzylmalonic acid derivative revealed an active site that discriminates prochiral carboxylates and accommodates the large size and distinct electrostatics of an α-carboxy substituent. This work emphasizes the potential of PylRS-derived enzymes for acylating tRNA with monomers whose α-substituent diverges substantially from the α-amine of proteinogenic amino acids. These enzymes or derivatives thereof could synergize with natural or evolved ribosomes and/or translation factors to generate diverse sequence-defined non-protein heteropolymers.