2024-04-30 韓国基礎科学研究院(IBS)
<関連情報>
- https://www.ibs.re.kr/cop/bbs/BBSMSTR_000000000738/selectBoardArticle.do
- https://www.nature.com/articles/s41467-024-47884-3
scLENS:偏りのないscRNA-seqデータ解析のためのデータ駆動型シグナル検出法 scLENS: data-driven signal detection for unbiased scRNA-seq data analysis
Hyun Kim,Won Chang,Seok Joo Chae,Jong-Eun Park,Minseok Seo & Jae Kyoung Kim
Nature Communications Published:27 April 2024
DOI:https://doi.org/10.1038/s41467-024-47884-3
Abstract
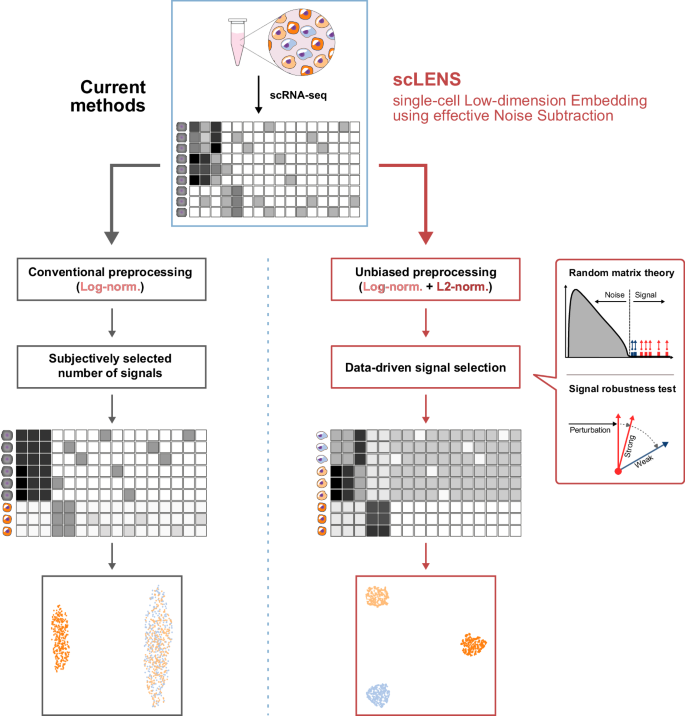
High dimensionality and noise have limited the new biological insights that can be discovered in scRNA-seq data. While dimensionality reduction tools have been developed to extract biological signals from the data, they often require manual determination of signal dimension, introducing user bias. Furthermore, a common data preprocessing method, log normalization, can unintentionally distort signals in the data. Here, we develop scLENS, a dimensionality reduction tool that circumvents the long-standing issues of signal distortion and manual input. Specifically, we identify the primary cause of signal distortion during log normalization and effectively address it by uniformizing cell vector lengths with L2 normalization. Furthermore, we utilize random matrix theory-based noise filtering and a signal robustness test to enable data-driven determination of the threshold for signal dimensions. Our method outperforms 11 widely used dimensionality reduction tools and performs particularly well for challenging scRNA-seq datasets with high sparsity and variability. To facilitate the use of scLENS, we provide a user-friendly package that automates accurate signal detection of scRNA-seq data without manual time-consuming tuning.