2024-09-18 ミュンヘン大学(LMU)
<関連情報>
- https://www.lmu.de/en/newsroom/news-overview/news/nanotechnology-dna-origami-with-cargo-function.html
- https://www.nature.com/articles/s41467-024-51721-y
- https://onlinelibrary.wiley.com/doi/10.1002/anie.202408295
DNA折り紙アレイにおけるアンチジャンクションの制御された機械化学的結合 Controlled mechanochemical coupling of anti-junctions in DNA origami arrays
Fiona Cole,Martina Pfeiffer,Dongfang Wang,Tim Schröder,Yonggang Ke & Philip Tinnefeld
Nature Communications Published:10 September 2024
DOI:https://doi.org/10.1038/s41467-024-51721-y
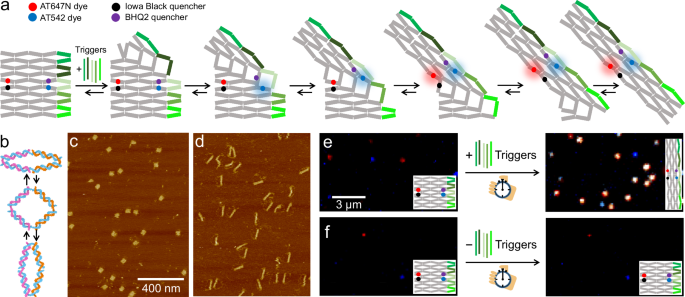
Abstract
Allostery is a hallmark of cellular function and important in every biological system. Still, we are only starting to mimic it in the laboratory. Here, we introduce an approach to study aspects of allostery in artificial systems. We use a DNA origami domino array structure which–upon binding of trigger DNA strands–undergoes a stepwise allosteric conformational change. Using two FRET probes placed at specific positions in the DNA origami, we zoom in into single steps of this reaction cascade. Most of the steps are strongly coupled temporally and occur simultaneously. Introduction of activation energy barriers between different intermediate states alters this coupling and induces a time delay. We then apply these approaches to release a cargo DNA strand at a predefined step in the reaction cascade to demonstrate the applicability of this concept in tunable cascades of mechanochemical coupling with both spatial and temporal control.
DNA折り紙小胞センサーとトリガーによる1分子貨物移動 DNA Origami Vesicle Sensors with Triggered Single-Molecule Cargo Transfer
Ece Büber, Renukka Yaadav, Tim Schröder, Henri G. Franquelim, Philip Tinnefeld
Angewandte Chemie International Edition Published: 09 September 2024
DOI:https://doi.org/10.1002/anie.202408295
Abstract
Interacting with living systems typically involves the ability to address lipid membranes of cellular systems. The first step of interaction of a nanorobot with a cell will thus be the detection of binding to a lipid membrane. Utilizing DNA origami, we engineered a biosensor with single-molecule Fluorescence Resonance Energy Transfer (smFRET) as transduction mechanism for precise lipid vesicle detection and cargo delivery. The system hinges on a hydrophobic ATTO647N modified single-stranded DNA (ssDNA) leash, protruding from a DNA origami nanostructure. In a vesicle-free environment, the ssDNA coils, ensuring high FRET efficiency. Upon vesicle binding to cholesterol anchors on the DNA origami, hydrophobic ATTO647N induces the ssDNA to stretch towards the lipid bilayer, reducing FRET efficiency. As the next step, the sensing strand serves as molecular cargo that can be transferred to the vesicle through a triggered strand displacement reaction. Depending on the number of cholesterols on the displacer strands, we either induce a diffusive release of the fluorescent load towards neighboring vesicles or a stoichiometric release of a single cargo-unit to the vesicle on the nanosensor. Ultimately, our multi-functional liposome interaction and detection platform opens up pathways for innovative biosensing applications and controllable stoichiometric loading of vesicles with single-molecule control.