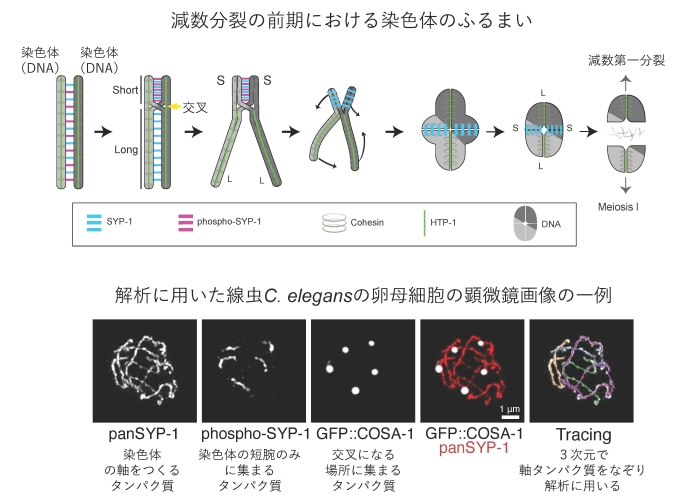
2024-10-11 コロンビア大学
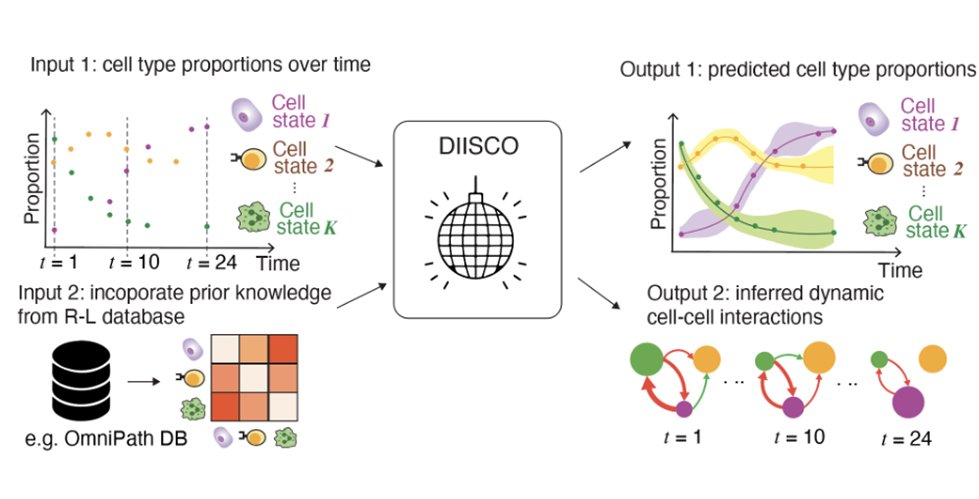 New Study Unveils DIISCO: A Revolutionary Method for Analyzing Dynamic Intercellular Interactions in Cancer. DIISCO is a machine learning method. Its input is cell-type proportions over time and gene expression of known receptor-ligand pairs. Its output is cell-type dynamics over time and networks characterizing the strength of cell-cell interactions and how they change over time. Image credit: Azizi Lab
New Study Unveils DIISCO: A Revolutionary Method for Analyzing Dynamic Intercellular Interactions in Cancer. DIISCO is a machine learning method. Its input is cell-type proportions over time and gene expression of known receptor-ligand pairs. Its output is cell-type dynamics over time and networks characterizing the strength of cell-cell interactions and how they change over time. Image credit: Azizi Lab
<関連情報>
- https://cancerdynamics.columbia.edu/news/new-study-unveils-diisco-revolutionary-method-analyzing-dynamic-intercellular-interactions
- https://genome.cshlp.org/content/early/2024/09/04/gr.279126.124.abstract
時系列の単一細胞データから動的な細胞間相互作用を推定するためのベイズフレームワーク A Bayesian framework for inferring dynamic intercellular interactions from time-series single-cell data
Cameron Y Park,Shouvik Mani,Nicolas Beltran-Velez,Katie Maurer,Teddy Huang,Shuqiang Li,Satyen Gohil,Kenneth J Livak,David A Knowles,Catherine J Wu andElham Azizi
Genome Research Published:September 5, 2024
DOI:10.1101/gr.279126.124
Abstract
Characterizing cell-cell communication and tracking its variability over time are crucial for understanding the coordination of biological processes mediating normal development, disease progression, and responses to perturbations such as therapies. Existing tools fail to capture time-dependent intercellular interactions, and primarily rely on existing databases compiled from limited contexts. We introduce DIISCO, a Bayesian framework designed to characterize the temporal dynamics of cellular interactions using single-cell RNA sequencing data from multiple time points. Our method utilizes structured Gaussian process regression to unveil time-resolved interactions among diverse cell types according to their coevolution and incorporates prior knowledge of receptor-ligand complexes. We show the interpretability of DIISCO in simulated data and new data collected from T cells co-cultured with lymphoma cells, demonstrating its potential to uncover dynamic cell-cell crosstalk.