2025-04-02 北京大学(PKU)
<関連情報>
- https://newsen.pku.edu.cn/news_events/news/research/14819.html
- https://www.nature.com/articles/s41467-025-57877-5
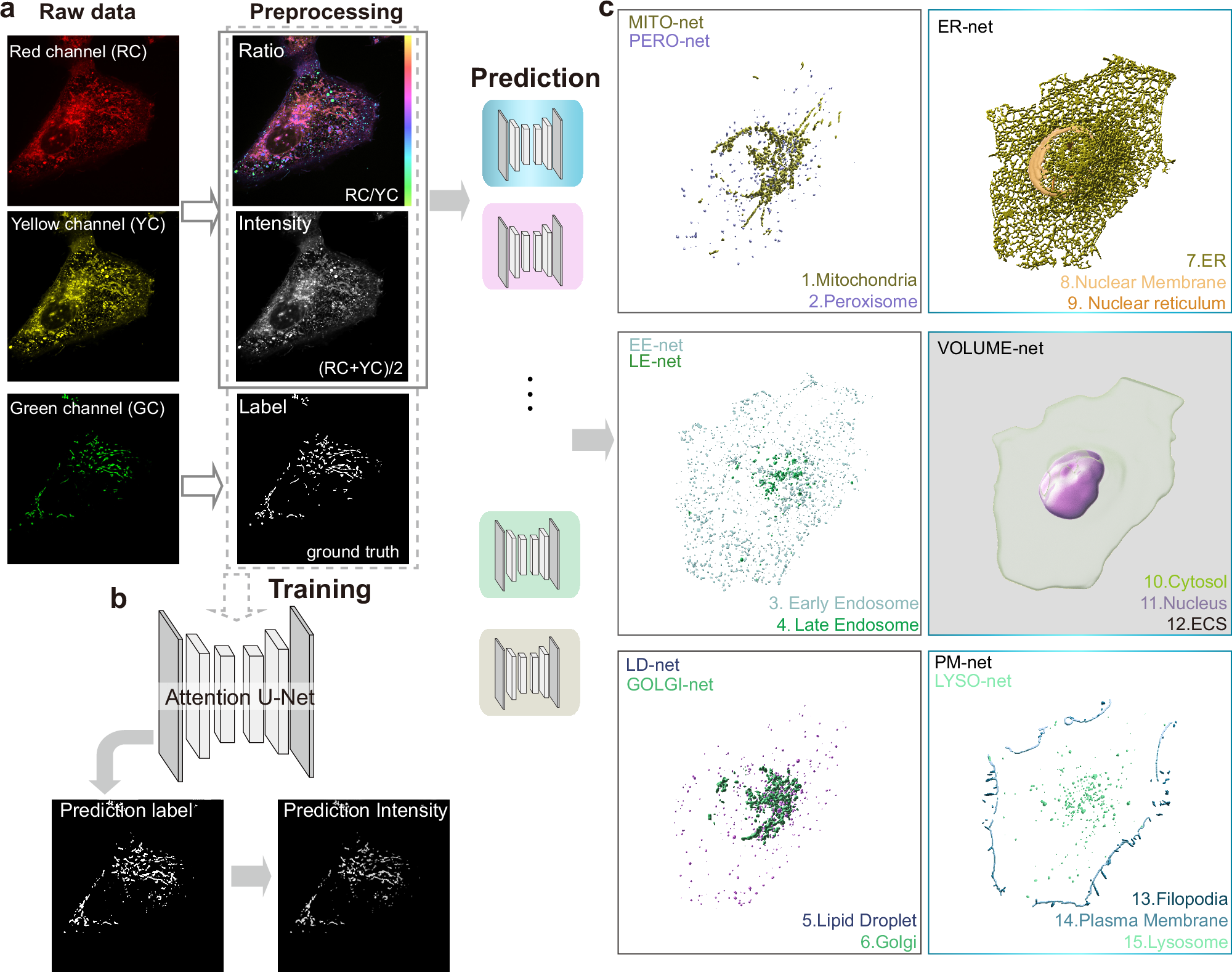
生きた細胞内の細胞小器官の高速セグメンテーションと多重イメージング Fast segmentation and multiplexing imaging of organelles in live cells
Karl Zhanghao,Meiqi Li,Xingye Chen,Wenhui Liu,Tianling Li,Yiming Wang,Fei Su,Zihan Wu,Chunyan Shan,Jiamin Wu,Yan Zhang,Jingyan Fu,Peng Xi & Dayong Jin
Nature Communications Published:21 March 2025
DOI:https://doi.org/10.1038/s41467-025-57877-5
Abstract
Studying organelles’ interactome at system level requires simultaneous observation of subcellular compartments and tracking their dynamics. Conventional multicolor approaches rely on specific fluorescence labeling, where the number of resolvable colors is far less than the types of organelles. Here, we use a lipid-specific dye to stain all the membrane-associated organelles and spinning-disk microscopes with an extended resolution of ~143 nm for high spatiotemporal acquisition. Due to the chromatic polarity sensitivity, high-resolution ratiometric images well reflect the heterogeneity of organelles. With deep convolutional neuronal networks, we successfully segmented up to 15 subcellular structures using one laser excitation. We further show that transfer learning can predict both 3D and 2D datasets from different microscopes, different cell types, and even complex systems of living tissues. We succeeded in resolving the 3D anatomic structure of live cells at different mitotic phases and tracking the fast dynamic interactions among six intracellular compartments with high robustness.


