2025-05-12 東京大学,理化学研究所,科学技術振興機構
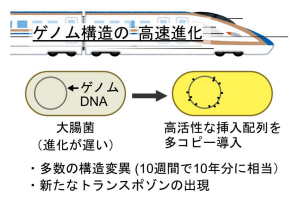
挿入配列(トランスポゾン)の導入による高速進化の概略
<関連情報>
- https://www.jst.go.jp/pr/announce/20250512/index.html
- https://www.jst.go.jp/pr/announce/20250512/pdf/20250512.pdf
- https://academic.oup.com/nar/article/53/9/gkaf331/8128223
挿入配列の活性化による細菌ゲノム構造の実験室進化 Laboratory evolution of the bacterial genome structure through insertion sequence activation
Yuki Kanai , Atsushi Shibai , Naomi Yokoi , Saburo Tsuru , Chikara Furusawa
Nucleic Acids Research Published:10 May 2025
DOI:https://doi.org/10.1093/nar/gkaf331
Abstract
The genome structure fundamentally shapes bacterial physiology, ecology, and evolution. Though insertion sequences (IS) are known drivers of drastic evolutionary changes in the genome structure, the process is typically slow and challenging to observe in the laboratory. Here, we developed a system to accelerate IS-mediated genome structure evolution by introducing multiple copies of a high-activity IS in Escherichia coli. We evolved the bacteria under relaxed neutral conditions, simulating those leading to IS expansion in host-restricted endosymbionts and pathogens. Strains accumulated a median of 24.5 IS insertions and underwent over 5% genome size changes within ten weeks, comparable to decades-long evolution in wild-type strains. The detected interplay of frequent small deletions and rare large duplications updates the view of genome reduction under relaxed selection from a simple consequence of the deletion bias to a nuanced picture including transient expansions. The high IS activity resulted in structural variants of IS and the emergence of composite transposons, illuminating potential evolutionary pathways for ISs and composite transposons. The extensive genome rearrangements we observed establish a baseline for assessing the fitness effects of IS insertions, genome size changes, and rearrangements, advancing our understanding of how mobile elements shape bacterial genomes.