2025-05-13 カリフォルニア大学サンタバーバラ校 (UCSB)
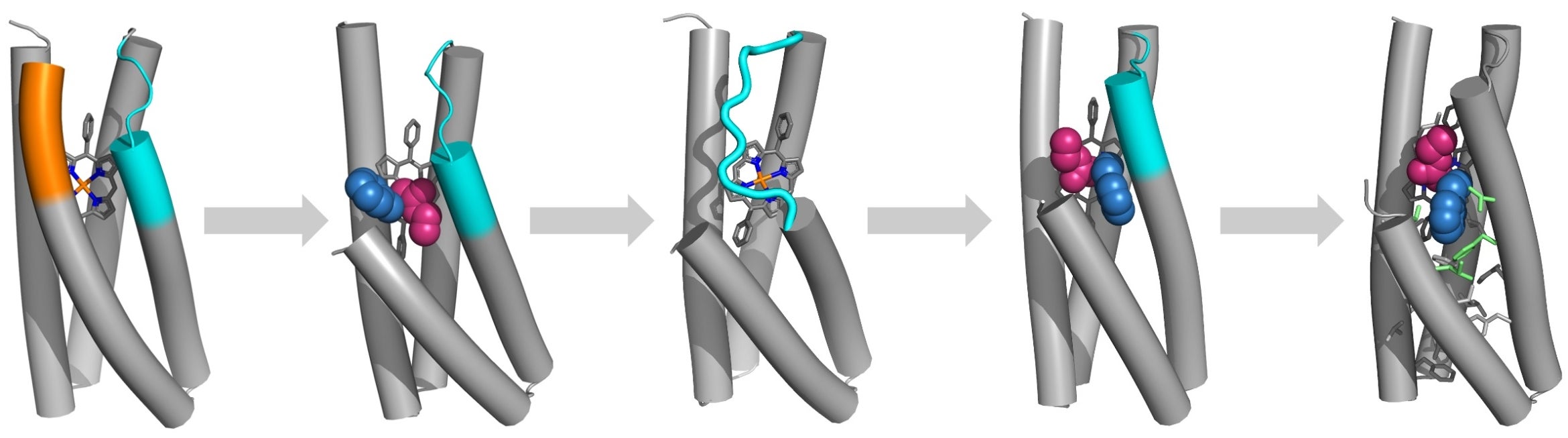 Photo Credit Courtesy image
Photo Credit Courtesy image
The researchers’ de novo protein design workflow begins with using a helical bundle as a framework for the new protein, left, using AI to design the amino acid sequences required for the desired protein structures, x-ray crystallography to assess the resulting protein structure, and further refinement to the design using tried-and-true methods and ‘chemical intuition,’ until the optimal design is reached, right.
<関連情報>
- https://news.ucsb.edu/2025/021875/enzymes-scratch
- https://www.science.org/doi/10.1126/science.adt7268
効率的かつ立体選択的な触媒としてのポルフィリン含有タンパク質の新規設計 De novo design of porphyrin-containing proteins as efficient and stereoselective catalysts
Kaipeng Hou, Wei Huang, Miao Qi, Thomas H. Tugwell, […] , and William F. DeGrado
Science Published:8 May 2025
Editor’s summary
One goal in protein design is the development of minimal scaffolds capable of retaining or improving the activities seen in larger proteins. Starting with a small, four-helix bundle protein that binds a heme derivative, Hou et al. redesigned the active site to enable facile substrate access while enhancing stereoselectivity and catalytic efficiency for styrene cyclopropanation. The authors also developed a version of the protein amenable to in-cell–directed evolution, enabling further optimization and expansion of the reactivity to silylation reactions. Their final design is highly active even in a mixed aqueous-organic solvent. —Michael A. Funk
Abstract
De novo design of protein catalysts with high efficiency and stereoselectivity provides an attractive approach toward the design of environmentally benign catalysts. Here, we design proteins that incorporate histidine-ligated synthetic porphyrin and heme ligands. Four of 10 designed proteins catalyzed cyclopropanation with an enantiomeric ratio greater than 99:1. A second class of proteins were designed to catalyze a silicon-hydrogen insertion and were optimized by directed evolution in whole cells. The evolved proteins incorporated features unlikely to be generated by computational design alone, including a proline in an α helix. Molecular dynamics simulations showed that as the proteins evolved toward higher activity, their conformational ensembles narrowed to favor more productive conformations. Our work demonstrates that efficient de novo protein catalysts are designable and should be useful for manifold chemical processes.