2025-07-16 米国国立標準技術研究所(NIST)
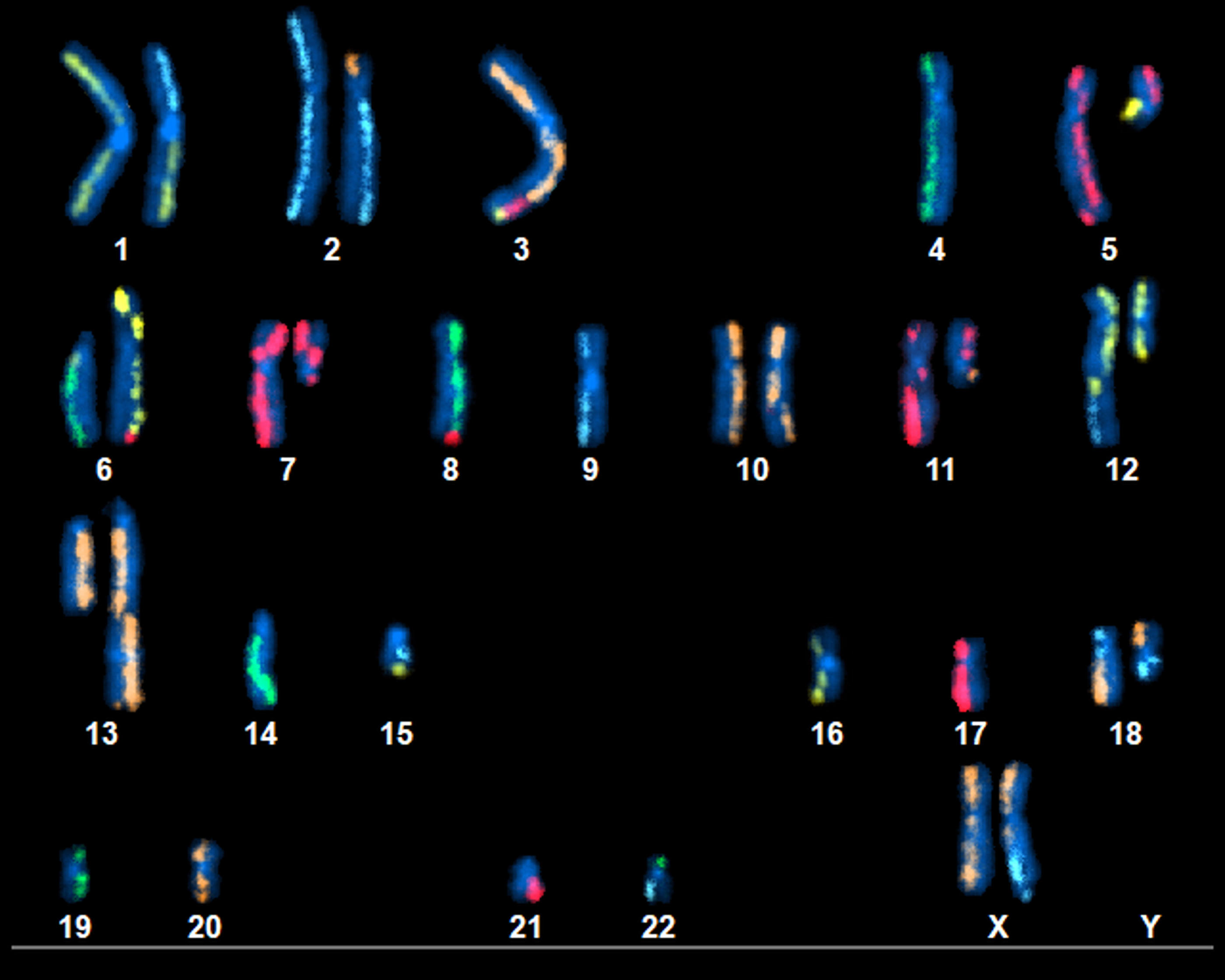
This image shows chromosomes in the pancreatic cancer cell line, where mixed colors indicate chromosomal rearrangements. For example, the first copy of chromosome 1 in the top left of the image is normal because both the top and bottom are yellow, but the top part of the second copy of chromosome 1 is replaced by the top part of chromosome 2 (blue) due to a rearrangement of chromosomes in the tumor. Image was created using the Kromatid KROMASURE Screen platform. Credit: NIST
<関連情報>
- https://www.nist.gov/news-events/news/2025/07/nist-releases-trove-genetic-data-spur-cancer-research
- https://www.nature.com/articles/s41597-025-05438-2
ボトルマッチングされた腫瘍と正常のペアにおける広範に同意されたゲノムの開発と広範な配列決定 Development and extensive sequencing of a broadly-consented Genome in a Bottle matched tumor-normal pair
Jennifer H. McDaniel,Vaidehi Patel,Nathan D. Olson,Hua-Jun He,Zhiyong He,Kenneth D. Cole,Alexander A. Gooden,Anthony Schmitt,Kristin Sikkink,Fritz J. Sedlazeck,Harsha Doddapaneni,Shalini N. Jhangiani,Donna M. Muzny,Marie-Claude Gingras,Heer Mehta,Sairam Behera,Luis F. Paulin,Alex R. Hastie,Hung-Chun Yu,Victor Weigman,Alison Rojas,Katie Kennedy,Jamie Remington,Isai Salas-González,… Justin M. Zook
Scientific Data Published:16 July 2025
DOI:https://doi.org/10.1038/s41597-025-05438-2
Abstract
The Genome in a Bottle Consortium (GIAB), hosted by the National Institute of Standards and Technology (NIST), is developing new matched tumor-normal samples, the first explicitly consented for public dissemination of genomic data and cell lines. Here, we describe a comprehensive genomic dataset from the first individual, HG008, including DNA from an adherent, epithelial-like pancreatic ductal adenocarcinoma (PDAC) tumor cell line and matched normal cells from duodenal and pancreatic tissues. Data for the tumor-normal matched samples comes from seventeen distinct state-of-the-art whole genome measurement technologies, including high depth short and long-read bulk whole genome sequencing (WGS), single cell WGS, Hi-C, and karyotyping. These data will be used by the GIAB Consortium to develop matched tumor-normal benchmarks for somatic variant detection. We expect these data to facilitate innovation for whole genome measurement technologies, de novo assembly of tumor and normal genomes, and bioinformatic tools to identify small and structural somatic variants. This first-of-its-kind broadly consented open-access resource will facilitate further understanding of sequencing methods used for cancer biology.