2025-08-08 ペンシルベニア州立大学(PennState)
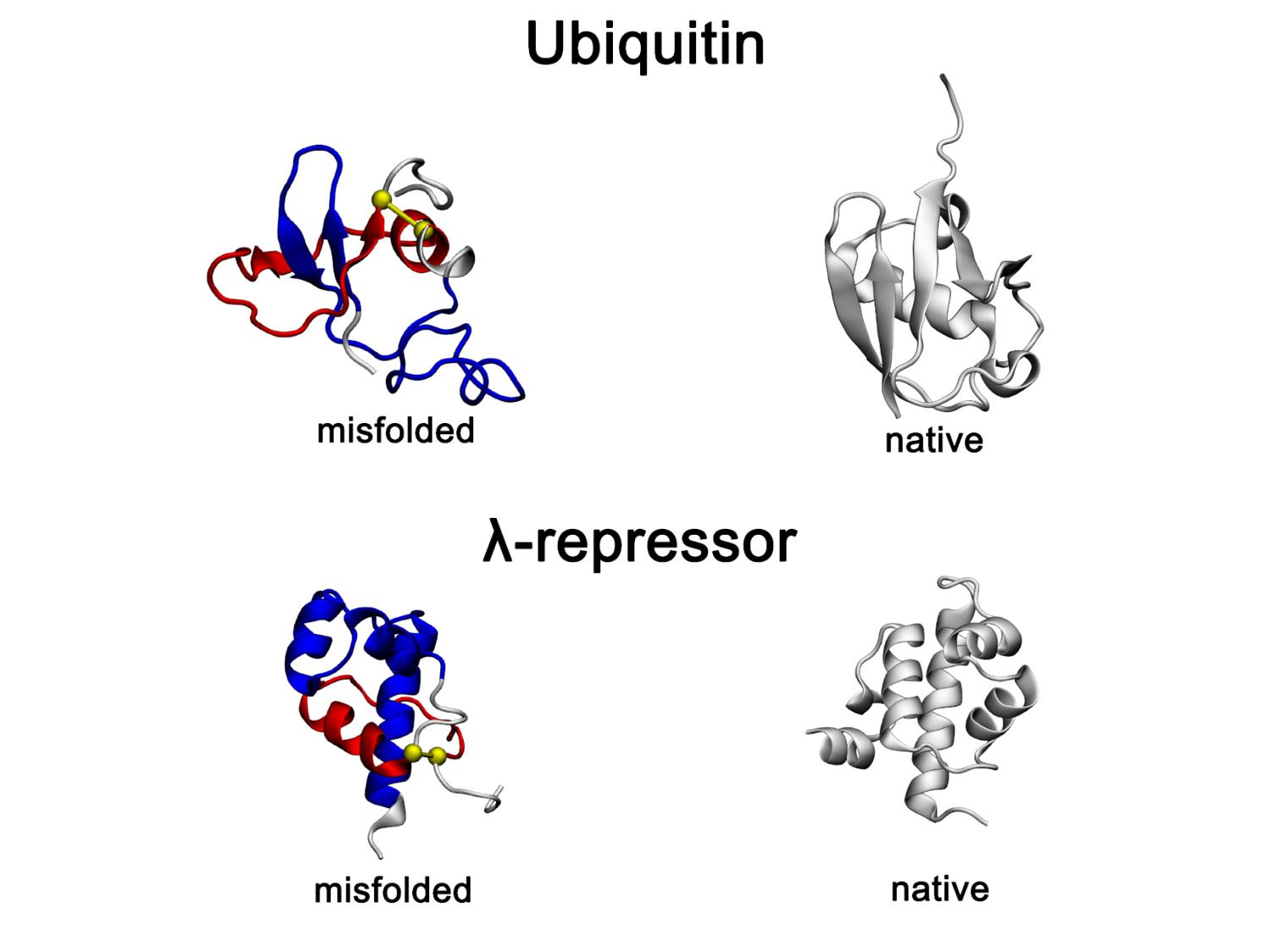
Recently identified and long-lasting type of protein misfolding — non-native entanglements — observed in all-atom protein folding simulations. Representative misfolded conformations of the small proteins, Ubiquitin and λ-repressor, exhibit gains-of-entanglement in all-atom folding simulations and are shown alongside their native structures. In the misfolded states, non-native entangled loops are highlighted in red, with yellow spheres marking loop closures and blue segments indicating threading through the loop. Credit: O’Brien Laboratory / Penn State. Creative Commons
<関連情報>
- https://www.psu.edu/news/eberly-college-science/story/new-class-protein-misfolding-simulated-high-definition
- https://www.science.org/doi/10.1126/sciadv.adt8974
非自生的な絡み合いタンパク質変性が全原子シミュレーションで観察され、実験的構造集合体によって支持されている Non-native entanglement protein misfolding observed in all-atom simulations and supported by experimental structural ensembles
Quyen V. Vu, Ian Sitarik, Yang Jiang, Yingzi Xia, […] , and Edward P. O’Brien
Science Advances Published:8 Aug 2025
DOI:https://doi.org/10.1126/sciadv.adt8974
Abstract
Several mechanisms are known to cause monomeric protein misfolding. Coarse-grained simulations have predicted an additional mechanism exists involving off-pathway, noncovalent lasso entanglements, which are long-lived kinetic traps and structurally resemble the native state. Here, we examine whether such misfolded states occur in long-timescale, all-atom folding simulations of ubiquitin and λ-repressor. We find that these entangled misfolded states are populated in higher-resolution models. However, because of the small size of ubiquitin and λ-repressor, these states are short-lived. In contrast, coarse-grained simulations of a larger protein, IspE, predict that it populates long-lived misfolded states. Using an Arrhenius extrapolation applied to all-atom simulations, we estimate that these IspE misfolded states have lifetimes similar to the native state while remaining soluble. We further show that these misfolded states are consistent with the structural changes inferred from limited proteolysis and cross-linking mass spectrometry experiments. Our results indicate that misfolded states composed of non-native entanglements can persist for long timescales in both all-atom simulations and experiments.

