2025-10-01 ミシガン大学
<関連情報>
- https://www.lsi.umich.edu/news/2025-10/catnip-chemists-new-data-driven-tool-broadens-access-greener-chemistry
- https://www.nature.com/articles/s41586-025-09519-5
化学物質とタンパク質の配列空間を結び付けて生体触媒反応を予測する Connecting chemical and protein sequence space to predict biocatalytic reactions
Alexandra E. Paton,Daniil A. Boiko,Jonathan C. Perkins,Nicholas I. Cemalovic,Thiago Reschützegger,Gabe Gomes & Alison R. H. Narayan
Nature Published:01 October 2025
DOI:https://doi.org/10.1038/s41586-025-09519-5
Abstract
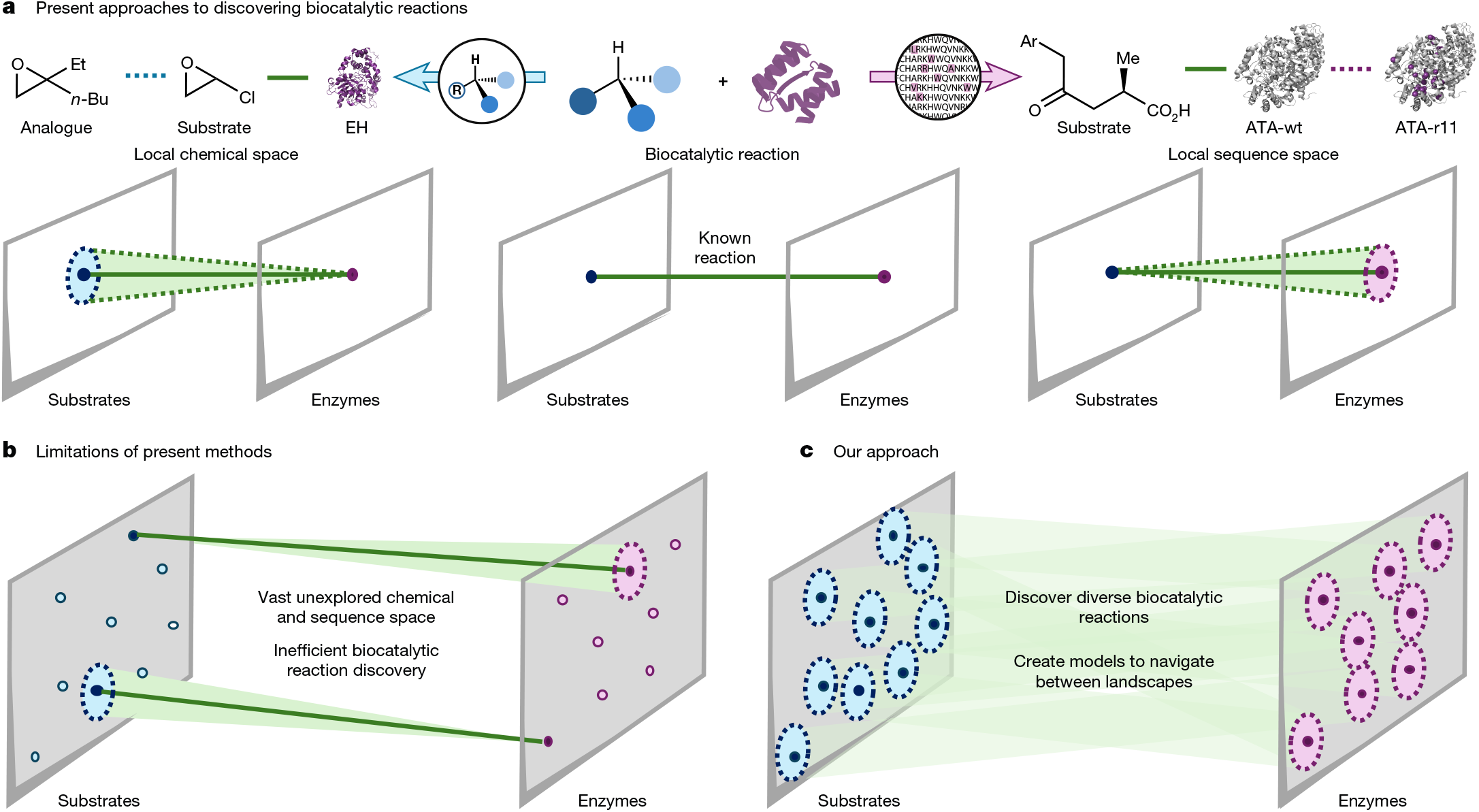
The application of biocatalysis in synthesis has the potential to offer streamlined routes towards target molecules1, tunable catalyst-controlled selectivity2, as well as processes with improved sustainability3. Despite these advantages, biocatalysis is often a high-risk strategy to implement, as identifying an enzyme capable of performing chemistry on a specific intermediate required for a synthesis can be a roadblock that requires extensive screening of enzymes and protein engineering to overcome4. Strategies for predicting which enzyme and small molecule are compatible have been hindered by the lack of well-studied biocatalytic reaction datasets5. The underexploration of connections between chemical and protein sequence space constrains navigation between these two landscapes. Here we report a two-phase effort relying on high-throughput experimentation to populate connections between productive substrate and enzyme pairs and the subsequent development of a tool, CATNIP, for predicting compatible α-ketoglutarate (α-KG)/Fe(ii)-dependent enzymes for a given substrate or, conversely, for ranking potential substrates for a given α-KG/Fe(ii)-dependent enzyme sequence. We anticipate that our approach can be readily expanded to further enzyme and transformation classes and will derisk the investigation and application of biocatalytic methods.