2026-01-26 カリフォルニア工科大学(Caltech)
<関連情報>
- https://www.caltech.edu/about/news/invention-dna-page-numbers-synthesis-kaihang-wang
- https://www.nature.com/articles/s41586-025-10006-0
DNA三元接合を用いた複雑かつ多様なDNA配列の構築 Construction of complex and diverse DNA sequences using DNA three-way junctions
Noah Evan Robinson,Weilin Zhang (张炜林),Rajesh Ghosh,Bryan Gerber,Hanqiao Zhang (张汉翘),Charles Sanfiorenzo,Sixiang Wang (王思翔),Dino Di Carlo & Kaihang Wang (王开航)
Nature Published:21 January 2026
DOI:https://doi.org/10.1038/s41586-025-10006-0
Abstract
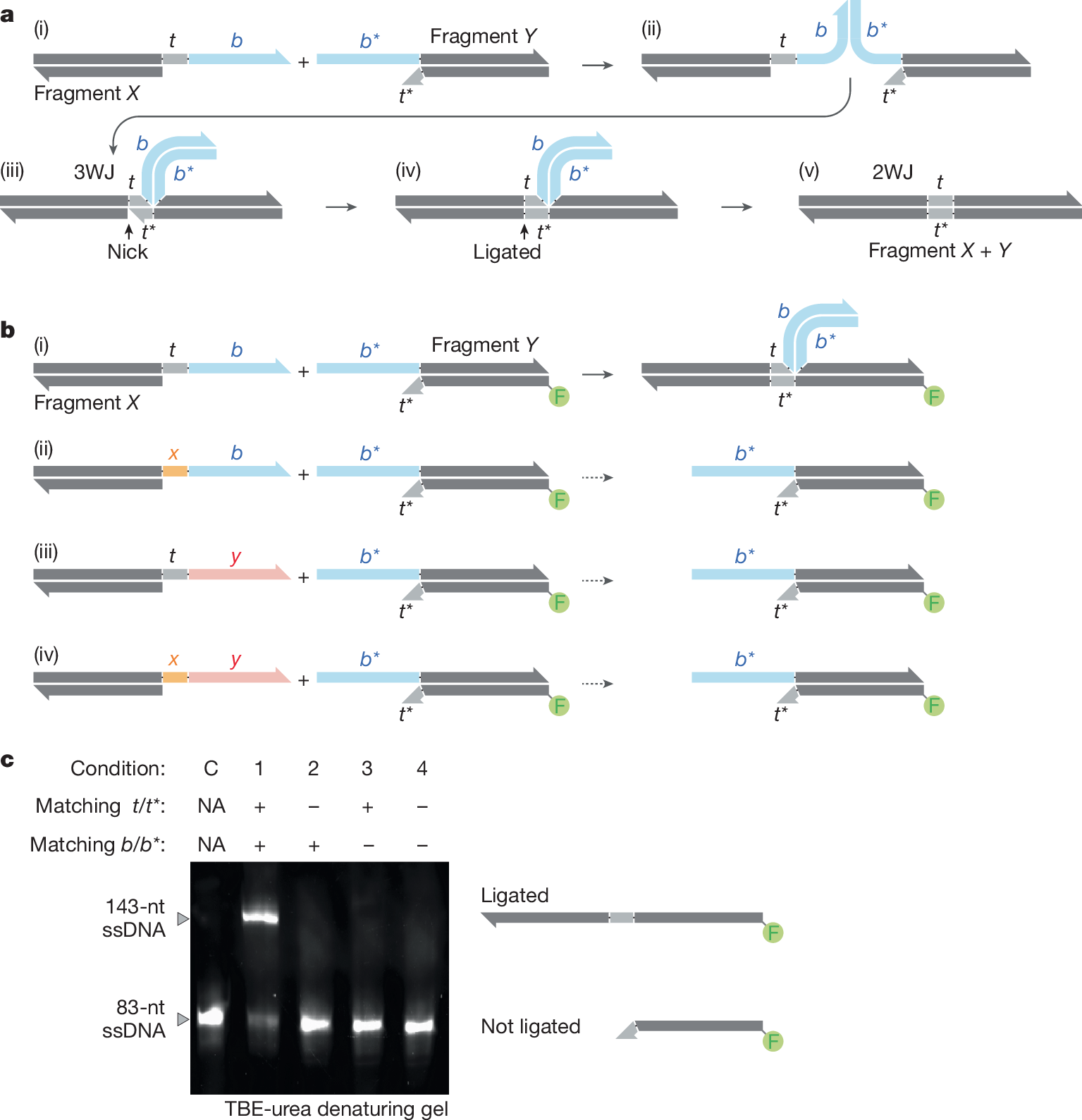
The ability to construct entirely new synthetic DNA sequences de novo is essential to engineering and studying biology. However, the ability to produce long complex synthetic DNA sequences and libraries currently lags behind the ability to sequence and edit DNA1,2. All existing DNA-assembly technologies rely on DNA sequence information found within the final construct to direct assembly between DNA molecules3,4,5,6,7,8,9,10,11. As a result of this paradigm, these sequences cannot be extensively optimized specifically for assembly without affecting the final sequence. To fundamentally address this challenge, here we show the development of a new DNA assembly technique named Sidewinder that separates the information that guides assembly from the final assembled sequence using DNA three-way junctions. We demonstrate the transformative nature of the Sidewinder technique with highly robust and accurate construction of a 40-piece multifragment assembly, complex DNA sequences of both high GC content and high repeats, parallel assembly of multiple distinct genes in the same reaction and a combinatorial library with a large number of diversified positions across the entire length of the gene for high coverage of a library of 442,368 variants. This technology enables high-fidelity DNA assembly with a misconnection rate at the three-way junction of approximately 1 in 1,000,000.