2026-02-13 東京大学
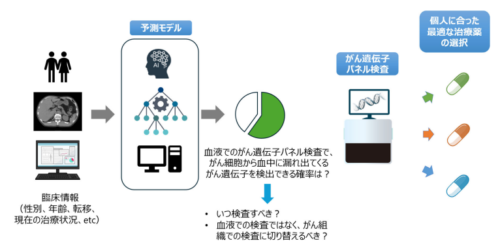
臨床情報をもとに、がん細胞の中のがん遺伝子を血液検査で検出できる確率を予測
<関連情報>
- https://www.h.u-tokyo.ac.jp/press/20260213-2.html
- https://www.h.u-tokyo.ac.jp/press/__icsFiles/afieldfile/2026/02/13/release_20260213.pdf
- https://www.sciencedirect.com/science/article/pii/S2059702926000116
日本の実世界データに基づく進行膵臓癌の液体包括的ゲノムプロファイリングにおけるctDNA検出能の予測モデル Prediction model for ctDNA detectability in liquid comprehensive genomic profiling of advanced pancreatic cancer based on Japanese real-world data
H. Ikushima, K. Watanabe, A. Shinozaki-Ushiku, K. Ishigaki, S. Kodera, N. Takeda, M. Fujishiro, K. Oda, H. Kage
ESMO Open Available online: 12 February 2026
DOI:https://doi.org/10.1016/j.esmoop.2026.106069
Highlights
- A prediction model for ctDNA detectability in advanced pancreatic cancer was developed.
- The model uses pre-test clinical information available before blood sampling.
- Liver/bone metastases predicted higher ctDNA detectability, while lung/peritoneal metastases predicted lower detectability.
- Model performance was validated in two independent nationwide test cohorts.
- The model may support optimal timing and modality selection for genomic profiling.
Background
Circulating tumor DNA (ctDNA) is sometimes undetectable in liquid comprehensive genomic profiling (CGP) of advanced-stage pancreatic cancer, resulting in false-negative findings that may mislead treatment decisions and waste health care resources. Predicting ctDNA detectability before testing may help optimize the timing of liquid CGP and guide decisions to consider alternative approaches, such as tissue-based CGP.
Patients and methods
We analyzed data from 7498 patients with advanced pancreatic adenocarcinoma using a nationwide real-world clinical genomic database. The tissue CGP cohort (cohort 1) included 4110 patients tested with FoundationOne CDx between June 2019 and December 2023. A prediction model for ctDNA detectability based on routinely available clinical variables before blood collection was trained on 2220 patients who underwent FoundationOne Liquid CDx between August 2021 and December 2023 (cohort 2). The model was deployed as a web application (https://pancreasliquidcgp.streamlit.app) and tested on two independent cohorts: 629 patients (cohort 3; FoundationOne Liquid CDx, January to December 2024) and 539 patients (cohort 4; Guardant360, July 2023 to December 2024). Model performance was assessed using Brier scores and calibration plots. Feature importance was evaluated using SHapley Additive exPlanations (SHAP).
Results
Among the 2220 patients in cohort 2, ctDNA was detected in 1130 (50.9%). The model achieved a Brier score of 0.210 and showed good calibration. SHAP analysis identified liver and bone metastases, progressive disease, and poor performance status as positive predictors of ctDNA detectability. Conversely, peritoneal and lung metastases negatively contributed to prediction. In the independent test cohorts, the model maintained robust performance with Brier scores of 0.203 (cohort 3) and 0.204 (cohort 4).
Conclusion
This prediction model enables accurate pre-test estimation of ctDNA detectability in advanced pancreatic cancer and may enhance the clinical utility of liquid CGP by informing optimal test selection and timing.