ストレス下でRNAの折り畳みが変化する様子を新たな手法で解明、最終的にはRNAが関与する疾患の治療法開発に役立つと期待される New method shows how RNA folding changes under stress and could eventually help in the development of treatment for RNA-linked diseases
2022-06-21 ペンシルベニア州立大学(PennState)
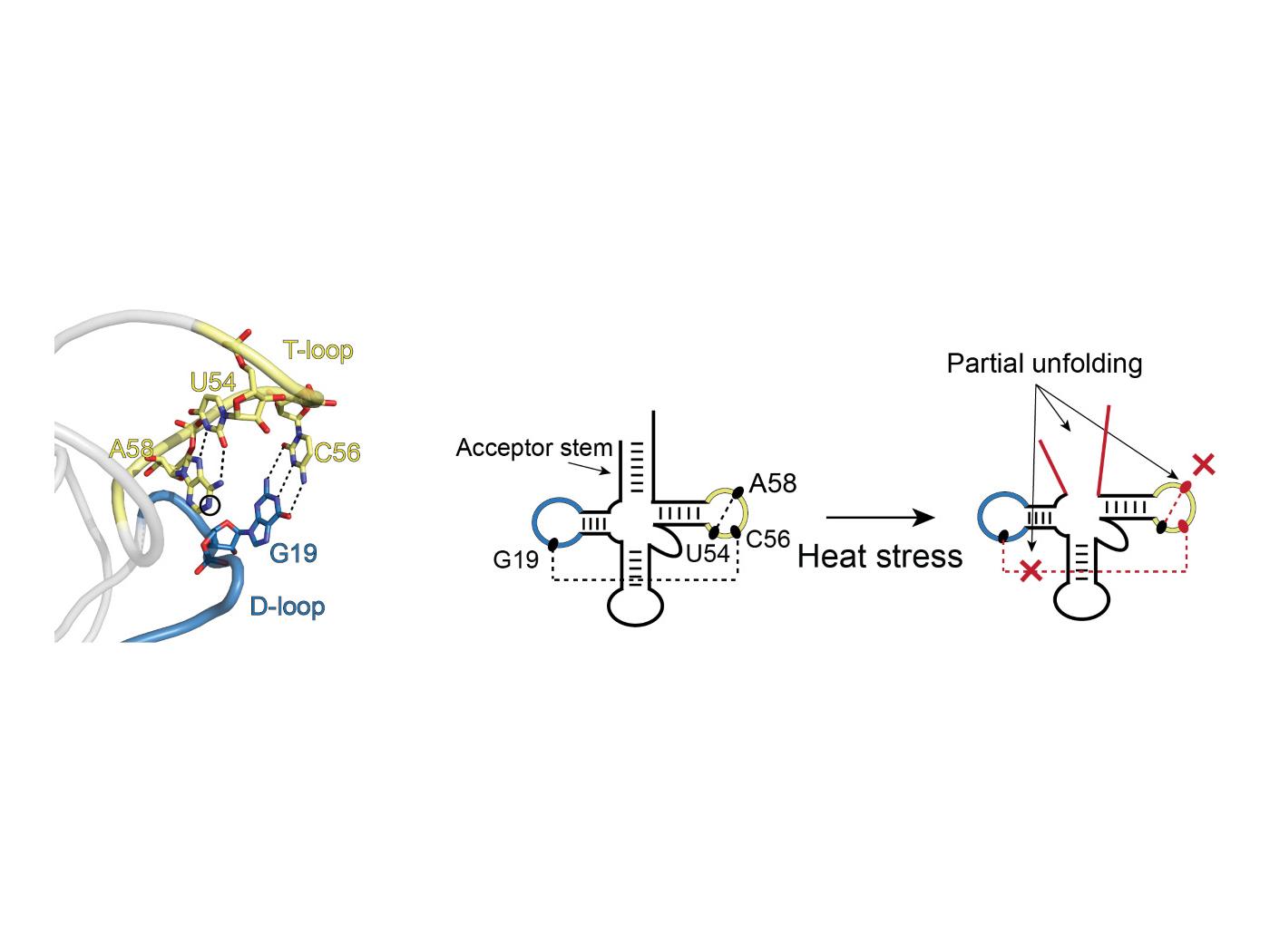 An example of partial unfolding of tRNA under heat stress. The image on the left shows the natural three-dimensional conformation of a portion of the tRNA with the D-loop (blue) and T-loop (yellow) connected in what is referred to as “kissing loops.” The two images on the right show the transition in structure that occurs upon heat stress. D- and T-loops retain their blue and yellow coloring. There is partial unfolding of the acceptor stem and the kissing loops no longer kiss. Credit: Bevilacqua Lab, Penn State. All Rights Reserved.
An example of partial unfolding of tRNA under heat stress. The image on the left shows the natural three-dimensional conformation of a portion of the tRNA with the D-loop (blue) and T-loop (yellow) connected in what is referred to as “kissing loops.” The two images on the right show the transition in structure that occurs upon heat stress. D- and T-loops retain their blue and yellow coloring. There is partial unfolding of the acceptor stem and the kissing loops no longer kiss. Credit: Bevilacqua Lab, Penn State. All Rights Reserved.
RNA分子の構造は、配列と、構造中に露出する特定のヌクレオチドに異なる化学タグを付加するジメチル硫酸(DMS)という薬品で分子を修飾することによって予測できる。
山上教授は、彼らの方法では、生きた細胞をDMSで処理した後、細胞からRNAを抽出し、サイズによって選別して、tRNAを単離すると説明しています。塩基配列を決定するためには、RNA分子のDNAコピーを作成する必要がある。そのためには、まずRNA分子の片端にプライマーと呼ばれる小さなDNA断片を結合させ、短いRNA配列の情報が失われるのを防ぐ。この酵素は、ある条件下で使用すると、ヌクレオチドに化学的修飾を加えても停止しない代わりに、修飾されたヌクレオチドになるとしばしばエラーを発生させるのだ。そこで研究者らは、このエラーを利用して、どのヌクレオチドが修飾されているかを特定し、「変異プロファイリング」と呼ばれるプロセスを経て、構造予測に役立てることにした。
この方法を用いれば、個々の分子のレベルで非常に正確な構造予測ができます。また、細胞内の各種tRNAの存在量や、タンパク質の効率的な生産と細胞全体の健康にとって重要な、いくつかの天然修飾の有無も明らかにすることができる。
<関連情報>
- https://www.psu.edu/news/eberly-college-science/story/determining-structure-small-rnas/
- https://www.pnas.org/doi/10.1073/pnas.2201237119
生体内tRNA構造体のゲノムワイド解析から、熱ストレス下でのRNA構造および修飾ダイナミクスを解明 Genome-wide analysis of the in vivo tRNA structurome reveals RNA structural and modification dynamics under heat stress
Ryota Yamagami, Jacob P. Sieg, Sarah M. Assmann and Philip C. Bevilacqua
Proceedings of the National Academy of Sciences Published:June 13, 2022
DOI:https://doi.org/10.1073/pnas.2201237119
Abstract
RNA structure plays roles in myriad cellular events including transcription, translation, and RNA processing. Genome-wide analyses of RNA secondary structure in vivo by chemical probing have revealed critical structural features of mRNAs and long ncRNAs. Here, we examine the in vivo secondary structure of a small RNA class, tRNAs. Study of tRNA structure is challenging because tRNAs are heavily modified and strongly structured. We introduce “tRNA structure-seq,” a new workflow that accurately determines in vivo secondary structures of tRNA. The workflow combines dimethyl sulfate (DMS) probing, ultra-processive RT, and mutational profiling (MaP), which provides mutations opposite DMS and natural modifications thereby allowing multiple modifications to be identified in a single read. We applied tRNA structure-seq to E. coli under control and stress conditions. A leading folding algorithm predicts tRNA structures with only ∼80% average accuracy from sequence alone. Strikingly, tRNA structure-seq, by providing experimental restraints, improves structure prediction under in vivo conditions to ∼95% accuracy, with more than 14 tRNAs predicted completely correctly. tRNA structure-seq also quantifies the relative levels of tRNAs and their natural modifications at single nucleotide resolution, as validated by LC-MS/MS. Our application of tRNA structure-seq yields insights into tRNA structure in living cells, revealing that it is not immutable but has dynamics, with partial unfolding of secondary and tertiary tRNA structure under heat stress that is correlated with a loss of tRNA abundance. This method is applicable to other small RNAs, including those with natural modifications and highly structured regions.