2023-05-12 ジョージア工科大学
◆この研究は、ジョージア工科大学とエモリー大学の生物医学工学のアーメット・コスクン教授の研究室から発表され、マルチプレックス免疫蛍光技術や機械学習を組み合わせた新しい方法を用いて、単一細胞におけるステム細胞治療の候補細胞の特定を可能にしました。
◆この研究は、患者のニーズに合わせて異なる場所から同じ細胞を収穫することが、特定の疾患の治療においてより良い結果をもたらす可能性があることを示しています。また、RNA分子の空間的な組織化を研究することで、細胞機能に重要な影響を与える隣接するRNA分子の多様性をより正確に分類することができることも明らかになりました。
<関連情報>
- https://research.gatech.edu/look-inside-stem-cells-helps-create-personalized-regenerative-medicine
- https://www.nature.com/articles/s41598-023-32474-y
- https://www.sciencedirect.com/science/article/pii/S2667237523000991?via%3Dihub
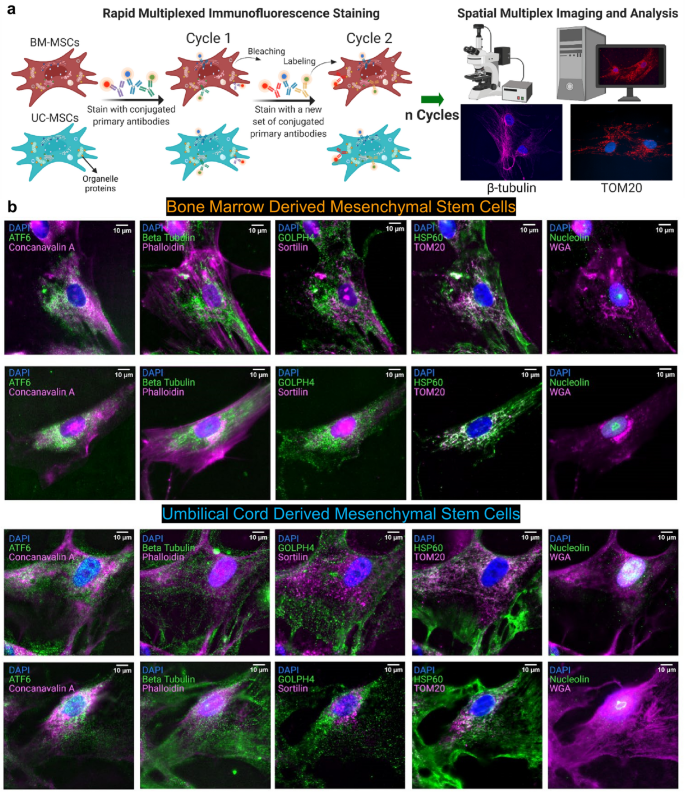
単一細胞内の空間的な細胞内オルガネラネットワーク Spatial subcellular organelle networks in single cells
Mythreye Venkatesan,Nicholas Zhang,Benoit Marteau,Yukina Yajima,Nerea Ortiz De Zarate Garcia,Zhou Fang,Thomas Hu,Shuangyi Cai,Adam Ford,Harrison Olszewski,Andrew Borst & Ahmet F. Coskun
Scientific Reports Published:01 April 2023
DOI:https://doi.org/10.1038/s41598-023-32474-y
Abstract
Organelles play important roles in human health and disease, such as maintaining homeostasis, regulating growth and aging, and generating energy. Organelle diversity in cells not only exists between cell types but also between individual cells. Therefore, studying the distribution of organelles at the single-cell level is important to understand cellular function. Mesenchymal stem cells are multipotent cells that have been explored as a therapeutic method for treating a variety of diseases. Studying how organelles are structured in these cells can answer questions about their characteristics and potential. Herein, rapid multiplexed immunofluorescence (RapMIF) was performed to understand the spatial organization of 10 organelle proteins and the interactions between them in the bone marrow (BM) and umbilical cord (UC) mesenchymal stem cells (MSCs). Spatial correlations, colocalization, clustering, statistical tests, texture, and morphological analyses were conducted at the single cell level, shedding light onto the interrelations between the organelles and comparisons of the two MSC subtypes. Such analytics toolsets indicated that UC MSCs exhibited higher organelle expression and spatially spread distribution of mitochondria accompanied by several other organelles compared to BM MSCs. This data-driven single-cell approach provided by rapid subcellular proteomic imaging enables personalized stem cell therapeutics.
単一細胞内における細胞内空間分解遺伝子近傍ネットワーク Subcellular spatially resolved gene neighborhood networks in single cells
Zhou Fang, Adam J. Ford, Thomas Hu, Nicholas Zhang, Athanasios Mantalaris, Ahmet F. Coskun
Cell Reports Methods Published: May 12, 2023
DOI:https://doi.org/10.1016/j.crmeth.2023.100476

Highlights
•spaGNN detects the hierarchy of gene proximity relationships
•Clustering algorithm identifies gene neighbors in subcellular patches
•Nearest neighborhood analysis reveals heterogeneous local gene proximity
•Gene proximity relationship outperforms gene expression in cell classification
Motivation
Spatially resolved transcriptomic technologies such as multiplexed error-robust fluorescence in situ hybridization (FISH) and sequential FISH generate high-dimensional datasets that reflect the organization of cells within tissues but also the organization of molecules within cells. We sought to develop an analysis method for inferring subcellular molecular interaction networks from image-based spatial omics data. This algorithm quantifies the physical proximity of RNA molecules when they are spatially located within the nearest neighborhood distances. Spatially resolved gene neighborhoods are used to generate networks that are distinct at different parts of a single cell. Decoding subcellular gene neighborhood networks enables downstream tasks such as cell classification better than overall gene expression per cell, thus expanding the use of subcellular spatial features for omics analysis.
Summary
Image-based spatial omics methods such as fluorescence in situ hybridization (FISH) generate molecular profiles of single cells at single-molecule resolution. Current spatial transcriptomics methods focus on the distribution of single genes. However, the spatial proximity of RNA transcripts can play an important role in cellular function. We demonstrate a spatially resolved gene neighborhood network (spaGNN) pipeline for the analysis of subcellular gene proximity relationships. In spaGNN, machine-learning-based clustering of subcellular spatial transcriptomics data yields subcellular density classes of multiplexed transcript features. The nearest-neighbor analysis produces heterogeneous gene proximity maps in distinct subcellular regions. We illustrate the cell-type-distinguishing capability of spaGNN using multiplexed error-robust FISH data of fibroblast and U2-OS cells and sequential FISH data of mesenchymal stem cells (MSCs), revealing tissue-source-specific MSC transcriptomics and spatial distribution characteristics. Overall, the spaGNN approach expands the spatial features that can be used for cell-type classification tasks.