2024-04-01 カリフォルニア大学サンディエゴ校(UCSD)
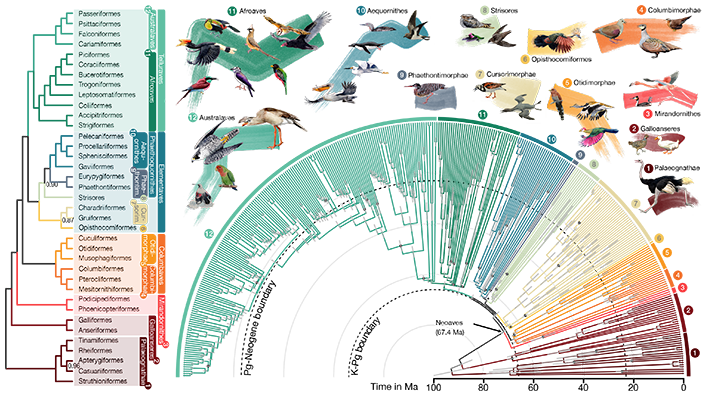
The updated bird family tree, published in Nature, delineating 93 million years of evolutionary relationships between 363 bird species. Credit: Jon Fjeldså (drawings) and Josefin Stiller
<関連情報>
- https://today.ucsd.edu/story/computational-tools-fuel-reconstruction-of-new-and-improved-bird-family-tree
- https://www.nature.com/articles/s41586-024-07323-1
- https://www.pnas.org/doi/10.1073/pnas.2319506121
鳥類進化の複雑さがファミリーレベルのゲノムから明らかになった Complexity of avian evolution revealed by family-level genomes
Josefin Stiller,Shaohong Feng,Al-Aabid Chowdhury,Iker Rivas-González,David A. Duchêne,Qi Fang,Yuan Deng,Alexey Kozlov,Alexandros Stamatakis,Santiago Claramunt,Jacqueline M. T. Nguyen,Simon Y. W. Ho,Brant C. Faircloth,Julia Haag,Peter Houde,Joel Cracraft,Metin Balaban,Uyen Mai,Guangji Chen,Rongsheng Gao,Chengran Zhou,Yulong Xie,Zijian Huang,Zhen Cao,… Guojie Zhang
Nature Published:01 April 2024
DOI:https://doi.org/10.1038/s41586-024-07323-1
We are providing an unedited version of this manuscript to give early access to its findings. Before final publication, the manuscript will undergo further editing. Please note there may be errors present which affect the content, and all legal disclaimers apply.
Abstract
Despite tremendous efforts in the past decades, relationships among main avian lineages remain heavily debated without a clear resolution. Discrepancies have been attributed to diversity of species sampled, phylogenetic method, and the choice of genomic regions 1–3. Here, we address these issues by analyzing genomes of 363 bird species 4 (218 taxonomic families, 92% of total). Using intergenic regions and coalescent methods, we present a well-supported tree but also a remarkable degree of discordance. The tree confirms that Neoaves experienced rapid radiation at or near the Cretaceous–Paleogene (K–Pg) boundary. Sufficient loci rather than extensive taxon sampling were more effective in resolving difficult nodes. Remaining recalcitrant nodes involve species that challenge modeling due to extreme GC content, variable substitution rates, incomplete lineage sorting, or complex evolutionary events such as ancient hybridization. Assessment of the impacts of different genomic partitions showed high heterogeneity across the genome. We discovered sharp increases in effective population size, substitution rates, and relative brain size following the K–Pg extinction event, supporting the hypothesis that emerging ecological opportunities catalyzed the diversification of modern birds. The resulting phylogenetic estimate offers novel insights into the rapid radiation of modern birds and provides a taxon-rich backbone tree for future comparative studies.
組換えが抑制された領域がネオアビ系統学を惑わす A region of suppressed recombination misleads neoavian phylogenomics
Siavash Mirarab, Iker Rivas-González, Shaohong Feng, +15, and Edward L. Braun
Proceedings of the National Academy of Sciences Published: April 1, 2024
DOI:https://doi.org/10.1073/pnas.2319506121