2024-11-21 ジョージア工科大学
<関連情報>
- https://research.gatech.edu/mapping-protein-interactions-fight-lung-cancer-coskun-pioneering-new-field-research
- https://www.nature.com/articles/s41467-023-43917-5
- https://medibio.tiisys.com/120480/
肺癌の薬物障害培養と組織における細胞内タンパク質間相互作用の空間分解解析 Spatially resolved subcellular protein–protein interactomics in drug-perturbed lung-cancer cultures and tissues
Shuangyi Cai,Thomas Hu,Abhijeet Venkataraman,Felix G. Rivera Moctezuma,Efe Ozturk,Nicholas Zhang,Mingshuang Wang,Tatenda Zvidzai,Sandip Das,Adithya Pillai,Frank Schneider,Suresh S. Ramalingam,You-Take Oh,Shi-Yong Sun & Ahmet F. Coskun
Nature Biomedical Engineering Published:30 October 2024
DOI:https://doi.org/10.1038/s41551-024-01271-x
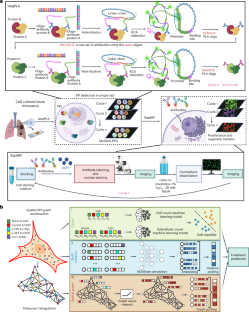
Abstract
Protein–protein interactions (PPIs) regulate signalling pathways and cell phenotypes, and the visualization of spatially resolved dynamics of PPIs would thus shed light on the activation and crosstalk of signalling networks. Here we report a method that leverages a sequential proximity ligation assay for the multiplexed profiling of PPIs with up to 47 proteins involved in multisignalling crosstalk pathways. We applied the method, followed by conventional immunofluorescence, to cell cultures and tissues of non-small-cell lung cancers with a mutated epidermal growth-factor receptor to determine the co-localization of PPIs in subcellular volumes and to reconstruct changes in the subcellular distributions of PPIs in response to perturbations by the tyrosine kinase inhibitor osimertinib. We also show that a graph convolutional network encoding spatially resolved PPIs can accurately predict the cell-treatment status of single cells. Multiplexed proximity ligation assays aided by graph-based deep learning can provide insights into the subcellular organization of PPIs towards the design of drugs for targeting the protein interactome.

00507-2/asset/89f5ad73-0424-4de0-85a8-8cca4e8e5393/main.assets/gr1.jpg)