2025-10-24 ペンシルベニア州立大学(PennState)
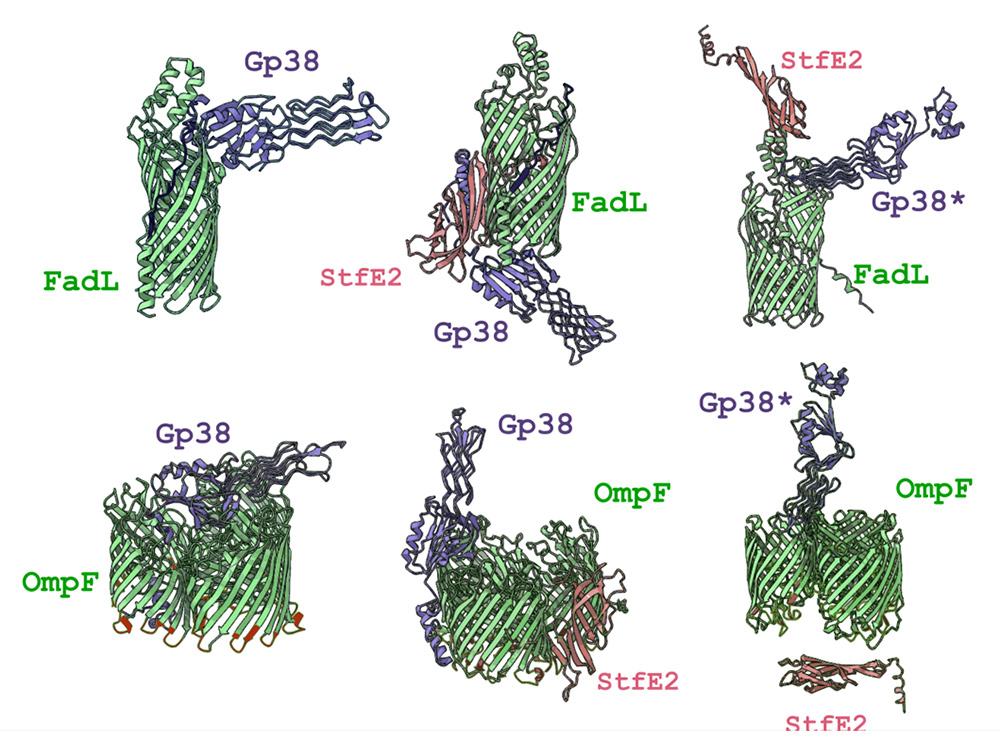
The proteins that allow viruses to land on bacteria — Gp38 — are attracted to FadL and OmpF, two proteins that help make up the outer walls of bacterial cells. StfE2, the chimeric protein formed with the help of the dormant prophage, stops the virus from landing on and infecting the bacteria. Credit: Provided by Thomas Wood. All Rights Reserved.
<関連情報>
- https://www.psu.edu/news/engineering/story/old-dog-new-tricks-prehistoric-viruses-can-be-used-defend-bacterial-cells
- https://academic.oup.com/nar/article/53/19/gkaf1041/8287591
ファージT2の吸着は、セリンリコンビナーゼPinQによる潜在性プロファージDNAの反転により阻害される Adsorption of phage T2 is inhibited due to inversion of cryptic prophage DNA by the serine recombinase PinQ
Joy Kirigo, Daniel Huelgas‐Méndez, María Tomás, Michael J Benedik, Rodolfo García‐Contreras, Thomas K Wood
Nucleic Acids Research Published:16 October 2025
DOI:https://doi.org/10.1093/nar/gkaf1041
Abstract
Recombinases catalyze site-specific integration, excision, and inversion of DNA and are often found adjacent to anti-phage system genes clustered in defense islands; however, their function in phage defense is unknown, as they are frequently dismissed as markers of prophages. Here, we characterize the physiological role of the previously uncharacterized serine recombinase PinQ (P segment inversion by Qin) of Escherichia coli cryptic prophage Qin and discover that it inhibits T2 phage infection by inverting a 1797 bp segment in a different cryptic prophage e14; this inversion leads to the formation of a novel protein from two chimeric genes, StfE2, that we find blocks phage adsorption. Modeling shows StfE2 inhibits T2 phage adsorption by preventing Gp38 binding to its primary receptors, porins FadL and OmpF. Corroborating the receptor-blocking hypothesis, T2 escape mutants evolve resistance to PinQ anti-phage defense by mutating gp38 in the hypervariable region 3. Therefore, we discovered the first recombinase-activated phage inhibition system.