2026-01-09 京都大学化学研究所
<関連情報>
- https://www.kuicr.kyoto-u.ac.jp/sites/topics/260109/
- https://www.kuicr.kyoto-u.ac.jp/sites/wp-content/uploads/ogata_20250109.pdf
- https://www.nature.com/articles/s41564-025-02234-x
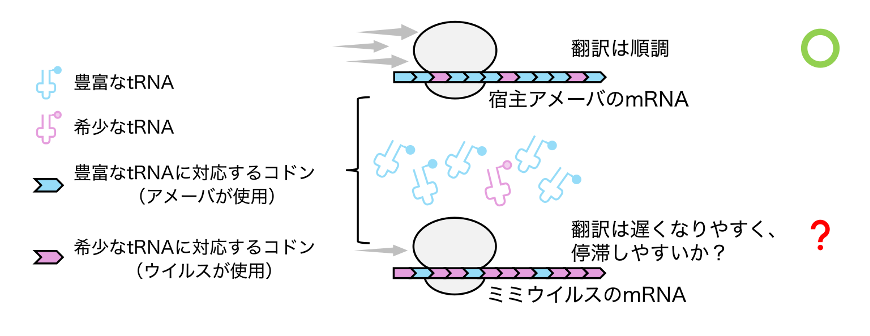
巨大ウイルスは、効率的な翻訳のためにアメーバ宿主内で特殊な細胞内環境を形成する A giant virus forms a specialized subcellular environment within its amoeba host for efficient translation
Ruixuan Zhang,Lotte Mayer,Hiroyuki Hikida,Yuichi Shichino,Mari Mito,Anouk Willemsen,Shintaro Iwasaki & Hiroyuki Ogata
Nature Microbiology Published:09 January 2026
DOI:https://doi.org/10.1038/s41564-025-02234-x
Abstract
Many eukaryotic viruses, including amoeba-infecting mimiviruses, have codon usage that deviates from their hosts. However, codon usage patterns that align with the cellular tRNA pool enable efficient translation. How these viruses cope with the mismatch between tRNA supply and demand is unclear. Here we show that Acanthamoeba polyphaga mimivirus (APMV) generates a subcellular area to translate viral mRNAs. tRNA sequencing showed that the tRNA pool was not substantially altered during the infection, even though the virus encodes tRNA genes. Using in situ labelling, we found that viral mRNAs and newly synthesized proteins were localized in the periphery of the viral factory, suggesting that APMV creates a discrete subcellular environment to facilitate translation. Frequently used codons in viral mRNAs had higher tRNA accessibility than the same type of codons in amoeba mRNAs. Our data show how local translation assists the virus in overcoming the mismatch between tRNA supply and demand.