2026-01-26 ノースカロライナ州立大学(NC State)
<関連情報>
- https://news.ncsu.edu/2026/01/environment-evolution-gut-microbiomes/
- https://link.springer.com/article/10.1186/s12862-025-02489-2
アフリカの草食動物群における腸内微生物叢の多様性に対する系統学的影響 Phylogenetic influence on gut microbiome diversity within an African herbivore community
Rylee Jensen,Erin A. McKenney,James C. Beasley,Claudine C. Cloete,Madeline Melton & Diana J. R. Lafferty
BMC Ecology and Evolution Published:20 December 2025
DOI:https://doi.org/10.1186/s12862-025-02489-2
Abstract
Background
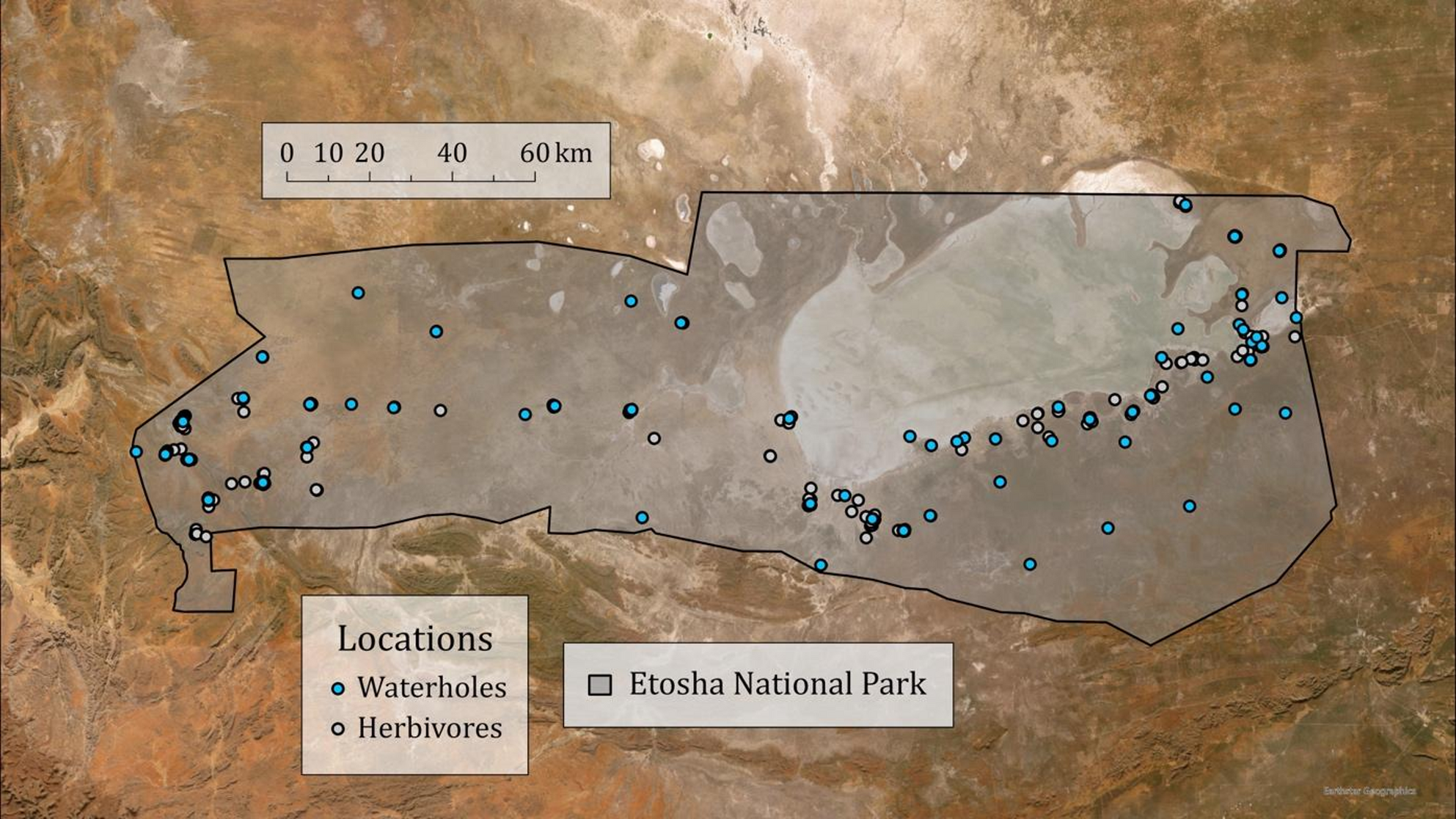
The microbial community within the gastrointestinal tract, known as the gut microbiome (GMB), is a complex micro-ecosystem that is modulated by the life history and physiological traits of the host as well as environmental conditions experienced by the host. In addition, phylogeny can be an important driver of GMB variability across mammalian species, with closely-related species sharing more similar microbial communities than distantly-related species, an eco-evolutionary pattern known as phylosymbiosis. In this study, we examined GMB diversity across 11 species of large herbivores in Etosha National Park (ENP), Namibia, to determine whether host species exhibit phylosymbiosis and whether different herbivore families host distinct microbial communities. The large herbivore community of ENP is an excellent model system because the herbivore species represent distinct evolutionary lineages and have evolved a variety of gut morphologies, dietary niches, and habitat requirements, all of which shape gut microbial diversity.
Results
While we found no evidence of phylosymbiosis across the greater ENP herbivore community, phylosymbiosis was detected among bovid species based on a positive correlation between microbial relative abundance and host evolutionary divergence times. Our results also revealed distinct microbial membership (e.g., Bacteroides, Treponema, and Alistipes) that distinguished bovid species from elephants and giraffes.
Conclusions
Our study provides new insights into the impact of phylogeny on GMB diversity in a closely-related African herbivore community. In particular, phylosymbiosis patterns observed in bovids but not all herbivore species demonstrates that microbial communities are dynamic and respond to a mixture of host evolutionary strategies and corresponding adaptations.