2023-08-03 エディンバラ大学
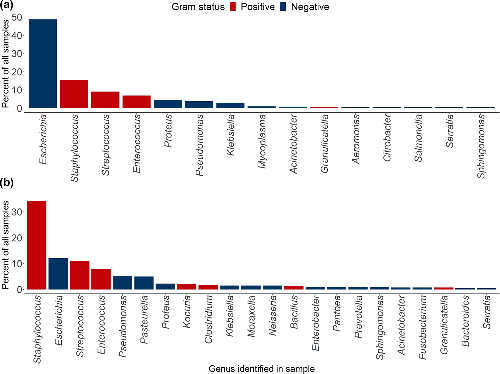
◆この手法は犬の一般的な細菌感染症を対象に開発され、細菌の同定にはメタゲノムDNA抽出とナノポアシーケンス技術を組み合わせています。これにより、数時間で細菌を同定し、適切な抗生物質の処方を可能としています。この新しい手法により、従来の培養法では同定が難しかった細菌種を同定し、抗生物質に対する耐性の可能性を高感度で判別することもできました。研究者たちは、このシステムをさまざまなサンプルや動物種の感染症に適応できるように設計しています。
◆さらに、将来的には、動物および人間の感染症の診断と治療に役立ち、ウイルスおよび寄生虫による他の種類の感染症にも応用できる可能性があると述べています。
<関連情報>
- https://www.ed.ac.uk/news/2023/rapid-infection-test-in-dogs-could-curb-antibiotic
- https://www.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.001066
犬の細菌感染症の診断と抗菌薬感受性予測のための迅速メタゲノムシークエンシング Rapid metagenomic sequencing for diagnosis and antimicrobial sensitivity prediction of canine bacterial infections
Natalie Ring, Alison S. Low, Bryan Wee, Gavin K. Paterson, Tim Nuttall, David Gally, Richard Mellanby, J. Ross Fitzgerald
Microbial Genomics Published: 20 July 2023
DOI: https://doi.org/10.1099/mgen.0.001066
ABSTRACT
Antimicrobial resistance is a major threat to human and animal health. There is an urgent need to ensure that antimicrobials are used appropriately to limit the emergence and impact of resistance. In the human and veterinary healthcare setting, traditional culture and antimicrobial sensitivity testing typically requires 48–72 h to identify appropriate antibiotics for treatment. In the meantime, broad-spectrum antimicrobials are often used, which may be ineffective or impact non-target commensal bacteria. Here, we present a rapid, culture-free, diagnostics pipeline, involving metagenomic nanopore sequencing directly from clinical urine and skin samples of dogs. We have planned this pipeline to be versatile and easily implementable in a clinical setting, with the potential for future adaptation to different sample types and animals. Using our approach, we can identify the bacterial pathogen present within 5 h, in some cases detecting species which are difficult to culture. For urine samples, we can predict antibiotic sensitivity with up to 95 % accuracy. Skin swabs usually have lower bacterial abundance and higher host DNA, confounding antibiotic sensitivity prediction; an additional host depletion step will likely be required during the processing of these, and other types of samples with high levels of host cell contamination. In summary, our pipeline represents an important step towards the design of individually tailored veterinary treatment plans on the same day as presentation, facilitating the effective use of antibiotics and promoting better antimicrobial stewardship.