2024-02-06 カリフォルニア大学サンディエゴ校(UCSD)
◆エンハンサー変異が生じると、例えばマウスや人間で指の追加が起こる可能性があり、この予測はリンクの強化によって不適切な遺伝子発現が引き起こされることが示唆された。Farley助教授らは、この知見を活かすことで、疾患の原因を特定し、遺伝子データを活用して精密医療を実現するための新たな可能性を開拓することを目指している。
<関連情報>
- https://today.ucsd.edu/story/extra-fingers-and-hearts-pinpointing-changes-to-our-genetic-instructions-that-disrupt-development
- https://www.nature.com/articles/s41586-023-06922-8
- https://www.sciencedirect.com/science/article/pii/S1534580723004926?via%3Dihub
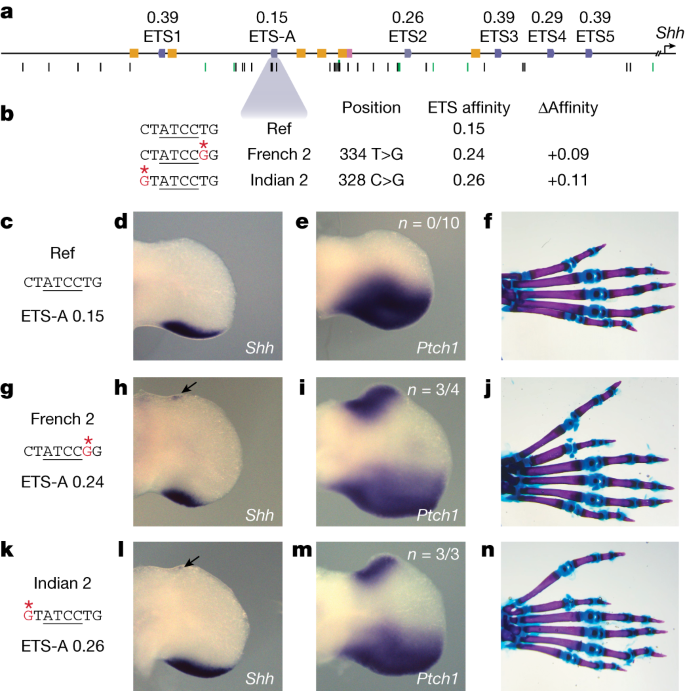
親和性を最適化するエンハンサー変異体が発達を阻害する Affinity-optimizing enhancer variants disrupt development
Fabian Lim,Joe J. Solvason,Genevieve E. Ryan,Sophia H. Le,Granton A. Jindal,Paige Steffen,Simran K. Jandu & Emma K. Farley
Nature Published:17 January 2024
DOI:https://doi.org/10.1038/s41586-023-06922-8
Abstract
Enhancers control the location and timing of gene expression and contain the majority of variants associated with disease1,2,3. The ZRS is arguably the most well-studied vertebrate enhancer and mediates the expression of Shh in the developing limb4. Thirty-one human single-nucleotide variants (SNVs) within the ZRS are associated with polydactyly4,5,6. However, how this enhancer encodes tissue-specific activity, and the mechanisms by which SNVs alter the number of digits, are poorly understood. Here we show that the ETS sites within the ZRS are low affinity, and identify a functional ETS site, ETS-A, with extremely low affinity. Two human SNVs and a synthetic variant optimize the binding affinity of ETS-A subtly from 15% to around 25% relative to the strongest ETS binding sequence, and cause polydactyly with the same penetrance and severity. A greater increase in affinity results in phenotypes that are more penetrant and more severe. Affinity-optimizing SNVs in other ETS sites in the ZRS, as well as in ETS, interferon regulatory factor (IRF), HOX and activator protein 1 (AP-1) sites within a wide variety of enhancers, cause gain-of-function gene expression. The prevalence of binding sites with suboptimal affinity in enhancers creates a vulnerability in genomes whereby SNVs that optimize affinity, even slightly, can be pathogenic. Searching for affinity-optimizing SNVs in genomes could provide a mechanistic approach to identify causal variants that underlie enhanceropathies.
心臓エンハンサー内の一塩基変異体が結合親和性を高め、心臓の発達を阻害することが明らかになった Single-nucleotide variants within heart enhancers increase binding affinity and disrupt heart development
Granton A. Jindal , Alexis T. Bantle , Joe J. Solvason , Jessica L. Grudzien , Agnieszka D’Antonio-Chronowska , Fabian Lim , Sophia H. Le , Benjamin P. Song , Michelle F. Ragsac , Adam Klie , Reid O. Larsen , Kelly A. Frazer , Emma K. Farley
Developmental Cell Published: October 16, 2023
DOI:https://doi.org/10.1016/j.devcel.2023.09.005
Highlights
•Low-affinity binding sites are prevalent within developmental heart enhancers
•Single-nucleotide variants that increase ETS affinity drive excess enhancer activity
•Affinity-optimizing enhancer variants disrupt heart development
Summary
Transcriptional enhancers direct precise gene expression patterns during development and harbor the majority of variants associated with phenotypic diversity, evolutionary adaptations, and disease. Pinpointing which enhancer variants contribute to changes in gene expression and phenotypes is a major challenge. Here, we find that suboptimal or low-affinity binding sites are necessary for precise gene expression during heart development. Single-nucleotide variants (SNVs) can optimize the affinity of ETS binding sites, causing gain-of-function (GOF) gene expression, cell migration defects, and phenotypes as severe as extra beating hearts in the marine chordate Ciona robusta. In human induced pluripotent stem cell (iPSC)-derived cardiomyocytes, a SNV within a human GATA4 enhancer increases ETS binding affinity and causes GOF enhancer activity. The prevalence of suboptimal-affinity sites within enhancers creates a vulnerability whereby affinity-optimizing SNVs can lead to GOF gene expression, changes in cellular identity, and organismal-level phenotypes that could contribute to the evolution of novel traits or diseases.
Graphical abstract