2024-06-26 ペンシルベニア州立大学(PennState)
<関連情報>
- https://www.psu.edu/news/research/story/researchers-compile-cacao-gene-atlas-help-plant-breeders-boost-chocolate-tree/
- https://link.springer.com/article/10.1186/s12870-024-05171-9
カカオ遺伝子アトラス:トランスクリプトーム発達アトラスはテオブロマ・カカオLにおける高度に組織特異的かつ動的に制御された遺伝子ネットワークを明らかにする The cacao gene atlas: a transcriptome developmental atlas reveals highly tissue-specific and dynamically-regulated gene networks in Theobroma cacao L
Evelyn Kulesza,Patrick Thomas,Sarah F. Prewitt,Akiva Shalit-Kaneh,Eric Wafula,Benjamin Knollenberg,Noah Winters,Eddi Esteban,Asher Pasha,Nicholas Provart,Craig Praul,Lena Landherr,Claude dePamphilis,Siela N. Maximova & Mark J. Guiltinan
BMC Plant Biology Published:26 June 2024
DOI:https://doi.org/10.1186/s12870-024-05171-9
Abstract
Background
Theobroma cacao, the cocoa tree, is a tropical crop grown for its highly valuable cocoa solids and fat which are the basis of a 200-billion-dollar annual chocolate industry. However, the long generation time and difficulties associated with breeding a tropical tree crop have limited the progress of breeders to develop high-yielding disease-resistant varieties. Development of marker-assisted breeding methods for cacao requires discovery of genomic regions and specific alleles of genes encoding important traits of interest. To accelerate gene discovery, we developed a gene atlas composed of a large dataset of replicated transcriptomes with the long-term goal of progressing breeding towards developing high-yielding elite varieties of cacao.
Results
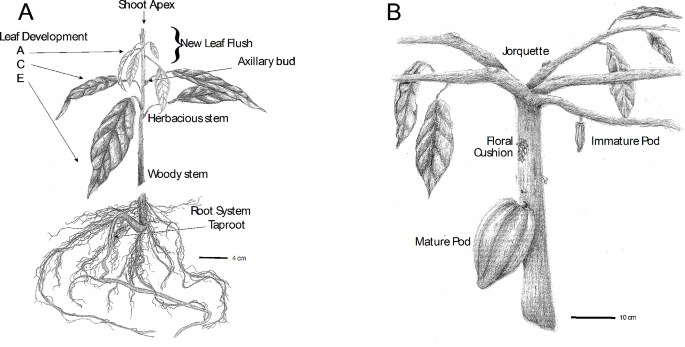
We describe the creation of the Cacao Transcriptome Atlas, its global characterization and define sets of genes co-regulated in highly organ- and temporally-specific manners. RNAs were extracted and transcriptomes sequenced from 123 different tissues and stages of development representing major organs and developmental stages of the cacao lifecycle. In addition, several experimental treatments and time courses were performed to measure gene expression in tissues responding to biotic and abiotic stressors. Samples were collected in replicates (3–5) to enable statistical analysis of gene expression levels for a total of 390 transcriptomes. To promote wide use of these data, all raw sequencing data, expression read mapping matrices, scripts, and other information used to create the resource are freely available online. We verified our atlas by analyzing the expression of genes with known functions and expression patterns in Arabidopsis (ACT7, LEA19, AGL16, TIP13, LHY, MYB2) and found their expression profiles to be generally similar between both species. We also successfully identified tissue-specific genes at two thresholds in many tissue types represented and a set of genes highly conserved across all tissues.
Conclusion
The Cacao Gene Atlas consists of a gene expression browser with graphical user interface and open access to raw sequencing data files as well as the unnormalized and CPM normalized read count data mapped to several cacao genomes. The gene atlas is a publicly available resource to allow rapid mining of cacao gene expression profiles. We hope this resource will be used to help accelerate the discovery of important genes for key cacao traits such as disease resistance and contribute to the breeding of elite varieties to help farmers increase yields.