2024-11-06 アルゴンヌ国立研究所(ANL)

Using the MProt-DPO framework, scientists created synthetic versions of malate dehydrogenase that preserve the protein’s critical structure and key binding areas. (Image by Arvind Ramanathan/Argonne National Laboratory.)
<関連情報>
- https://www.anl.gov/article/argonne-team-breaks-new-ground-in-aidriven-protein-design
- https://sc24.conference-program.com/presentation/?id=gb101&sess=sess497
- https://arxiv.org/abs/2302.04611
MProt-DPO:直接選好最適化により、マルチモーダルなタンパク質設計ワークフローのエクサフロップスの壁を破る MProt-DPO: Breaking the ExaFLOPS Barrier for Multimodal Protein Design Workflows with Direct Preference Optimization
Gautham Dharuman,Kyle Hippe,,Alexander Brace,Sam Foreman,Varuni Sastry,Huihuo Zheng,Logan Ward,Archit Vasan,Bharat Kale,Carla Mann,Heng Ma,Yun-Hsuan Cheng,Shengchao Liu,Chaowei Xiao,Koichi Yamada,Intel Corporation,Murali Emani,Thomas Gibbs,Ian Foster,Rick Stevens,Anima Anandkumar,Venkatram Vishwanath,Arvind Ramanathan,Yuliana Zamora,Servesh Muralidharan,Michael E. Papka
SessionACM Gordon Bell Finalists Presentations 2
DescriptionWe present a scalable, end-to-end workflow for protein design. By augmenting protein sequences with natural language descriptions of biochemical properties, we train generative models to preferentially align with protein fitness landscapes. Through complex experimental- and simulation-based observations, we integrate these measures as preferred parameters for generating new protein variants and demonstrate our workflow on five diverse supercomputers. We achieve >1 ExaFLOPS sustained performance in mixed precision on each supercomputer and a maximum sustained performance of 4.11 ExaFLOPS and peak performance of 5.57 ExaFLOPS. We establish the performance of our model on two tasks: (1) across a predetermined benchmark dataset of deep mutational scanning experiments to optimize the fitness-determining mutations in the yeast protein HIS7, and (2) in optimizing the design of the enzyme malate dehydrogenase to achieve lower activation barriers (and therefore increased catalytic rates) using simulation data. Our implementation thus sets high watermarks for multimodal protein design workflows.
テキスト誘導型タンパク質設計フレームワーク A Text-guided Protein Design Framework
Shengchao Liu, Yanjing Li, Zhuoxinran Li, Anthony Gitter, Yutao Zhu, Jiarui Lu, Zhao Xu, Weili Nie, Arvind Ramanathan, Chaowei Xiao, Jian Tang, Hongyu Guo, Anima Anandkumar
ArXix last revised 12 Aug 2024 (this version, v3
DOI:https://doi.org/10.48550/arXiv.2302.04611
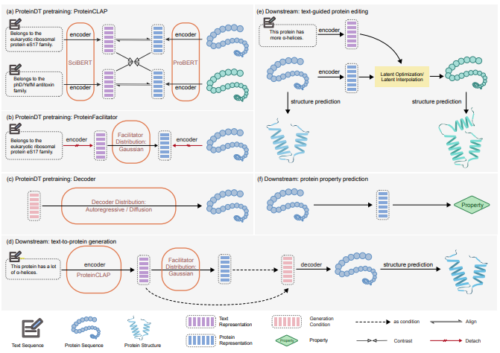
Abstract
Current AI-assisted protein design mainly utilizes protein sequential and structural information. Meanwhile, there exists tremendous knowledge curated by humans in the text format describing proteins’ high-level functionalities. Yet, whether the incorporation of such text data can help protein design tasks has not been explored. To bridge this gap, we propose ProteinDT, a multi-modal framework that leverages textual descriptions for protein design. ProteinDT consists of three subsequent steps: ProteinCLAP which aligns the representation of two modalities, a facilitator that generates the protein representation from the text modality, and a decoder that creates the protein sequences from the representation. To train ProteinDT, we construct a large dataset, SwissProtCLAP, with 441K text and protein pairs. We quantitatively verify the effectiveness of ProteinDT on three challenging tasks: (1) over 90\% accuracy for text-guided protein generation; (2) best hit ratio on 12 zero-shot text-guided protein editing tasks; (3) superior performance on four out of six protein property prediction benchmarks.