2025-03-01 中国科学院 (CAS)
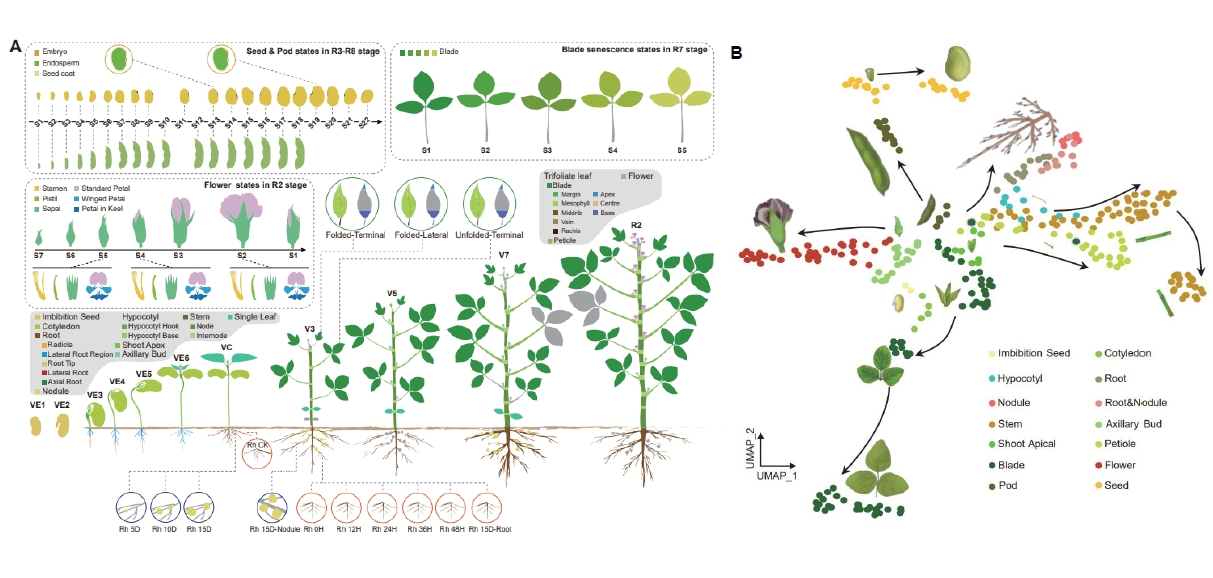
The transcriptome map of soybean organ development. (Image by IGDB)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202503/t20250303_902827.shtml
- https://www.sciencedirect.com/science/article/pii/S1674205225000693
ダイズ器官発生のための大規模統合トランスクリプトームアトラス A large-scale integrated transcriptomic atlas for soybean organ development
Jingwei Fan, Yanting Shen, Chuan Chen, Xi Chen, Xiaoyue Yang, Haixia Liu, Ruiying Chen, Shulin Liu, Bohan Zhang, Min Zhang, Guoan Zhou, Yu Wang, Haixi Sun, Yuqiang Jiang, Xiaofeng Wei, Tao Yang, Yucheng Liu, Dongmei Tian, Ziqing Deng, Xun Xu, Xin Liu, Zhixi Tian
Molecular Plant Published:Available online: 18 February 2025
DOI:https://doi.org/10.1016/j.molp.2025.02.003
SUMMARY
Soybean is one of the most important crops in the world and its production needs to be significantly increased to meet the escalating global demand. Elucidating the genetic regulatory networks underlying soybean organ development is critical for breeding elite and resilient varieties to ensure an increase in soybean production under the changing climates. Integrated transcriptomic atlas that leverages multiple types of transcriptomic data can facilitate the characterization of temporal-spatial expression patterns of most organ development-related genes and thereby help understand organ developmental processes. Here, we constructed a comprehensive integrated transcriptomic atlas for soybean, integrating bulk RNA-seq dataset from 314 samples across the soybean life cycle, along with snRNA-seq and Stereo-seq datasets from five organs: root, nodule, shoot apical, leaf and stem. Taking the investigations of genes related to organ specificity, blade development and nodule formation as examples, we show that the atlas has robust power for exploring key genes involved in organ formation. In addition, we built a user-friendly panoramic database for the transcriptomic atlas, facilitating easy access and queries, which will serve as a valuable resource to significantly advance future soybean functional studies.