2025-04-14 バーミンガム大学
<関連情報>
- https://www.birmingham.ac.uk/news/2025/light-bulb-moment-for-understanding-dna-repair-switches
- https://www.nature.com/articles/s41467-025-56974-9
- https://www.cell.com/molecular-cell/fulltext/S1097-2765(25)00109-1?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS1097276525001091%3Fshowall%3Dtrue
PIN1-SUMO2/3モチーフはRNF168の過剰なクロマチン蓄積とユビキチンシグナル伝達を抑制しIR抵抗性を促進する PIN1-SUMO2/3 motif suppresses excessive RNF168 chromatin accumulation and ubiquitin signaling to promote IR resistance
Anoop S. Chauhan,Matthew J. W. Mackintosh,Joseph Cassar,Alexander J. Lanz,Mohammed Jamshad,Hannah L. Mackay,Alexander J. Garvin,Alexandra K. Walker,Satpal S. Jhujh,Teresa Carlomagno,Aneika C. Leney,Grant S. Stewart & Joanna R. Morris
Nature Communications Published:14 April 2025
DOI:https://doi.org/10.1038/s41467-025-56974-9
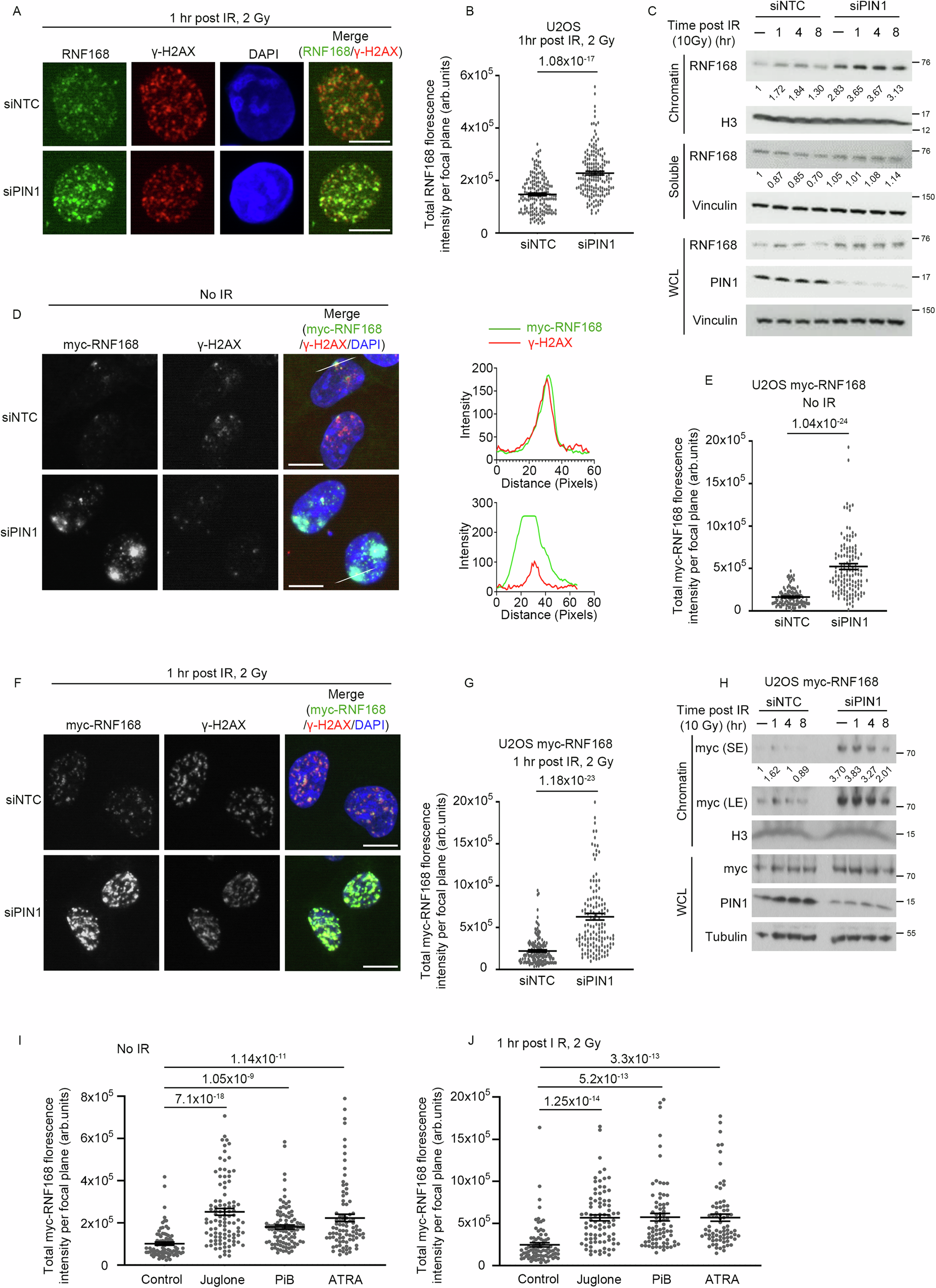
Abstract
RNF168 is an E3 ubiquitin ligase critical to the mammalian DNA double-strand break repair response. The protein is recruited to and amplifies ubiquitin signals at damaged chromatin and, if not properly regulated, can drive an uncontrolled ubiquitin cascade potentially harmful to repair outcomes. Several indirect mechanisms restrict RNF168 positive feedback, and a longstanding question has been whether these alone suppress excessive RNF168 signaling or whether mechanisms to remove RNF168 from damaged chromatin exist. Here, we reveal a cascade of post-translational modifications which act at three adjacent amino acids, threonine-208, proline-209 and lysine-210, to process RNF168 actively. Phosphorylation at threonine-208 by CDK1/2 induces interaction with the peptidyl-prolyl isomerase PIN1. PIN1 promotes RNF168 SUMOylation at lysine-210, resulting in p97/VCP mediated removal. These actions promote RNF168 clearance and limit RNF168 chromatin build-up. Thus, single amino acid substitutions of the regulatory motif (SUMO-PIN1-assisted Chromatin Regulator, SPaCR) that restrict PIN1 interaction or SUMOylation are sufficient to drive supraphysiological accumulation of RNF168, increased ubiquitin signaling, excessive 53BP1 recruitment and radiosensitivity. Our findings define a mechanism of direct RNF168 regulation that is part of the normal damage response, promoting RNF168 dissociation from chromatin and limiting deleterious ubiquitin signaling.
SUMO4はDNA二本鎖切断修復に必要なSUMO脱共役を促進する SUMO4 promotes SUMO deconjugation required for DNA double-strand-break repair
Alexander J. Garvin∙ Alexander J. Lanz∙ George E. Ronson∙ … ∙ Jai S. Bhachoo∙ Aneika C. Leney∙ Joanna R. Morris
Molecular Cell Accepted: February 5, 2025
DOI:https://doi.org/10.1016/j.molcel.2025.02.004
Graphical abstract

Highlights
- The roles of the SUMO family members, SUMO1–4, in DSB repair are not redundant
- SUMO4 conjugation is deleterious
- SUMO4 promotes SENP1 catalytic activity
- SUMO4:SENP1 restrict SUMO signaling and accumulation of the BRCA1-A complex
Summary
The amplitudes of small-modifier protein signaling through ubiquitin and the small ubiquitin-like modifiers, SUMO1–3, are critical to the correct phasing of DNA repair protein accumulation, activity, and clearance and for the completion of mammalian DNA double-strand-break (DSB) repair. However, how SUMO-conjugate signaling in the response is delineated is poorly understood. At the same time, the role of the non-conjugated SUMO protein, SUMO4, has remained enigmatic. Here, we reveal that human SUMO4 is required to prevent excessive DNA-damage-induced SUMOylation and deleterious over-accumulation of RAP80. Mechanistically we show that SUMO4 acts independently of its conjugation and potentiates SENP1 catalytic activity. These data identify SUMO4 as a SUMO deconjugation component and show that SUMO4:SENP1 are critical regulators of DNA-damage-induced SUMO signaling.

