2025-06-12 中国科学院(CAS)
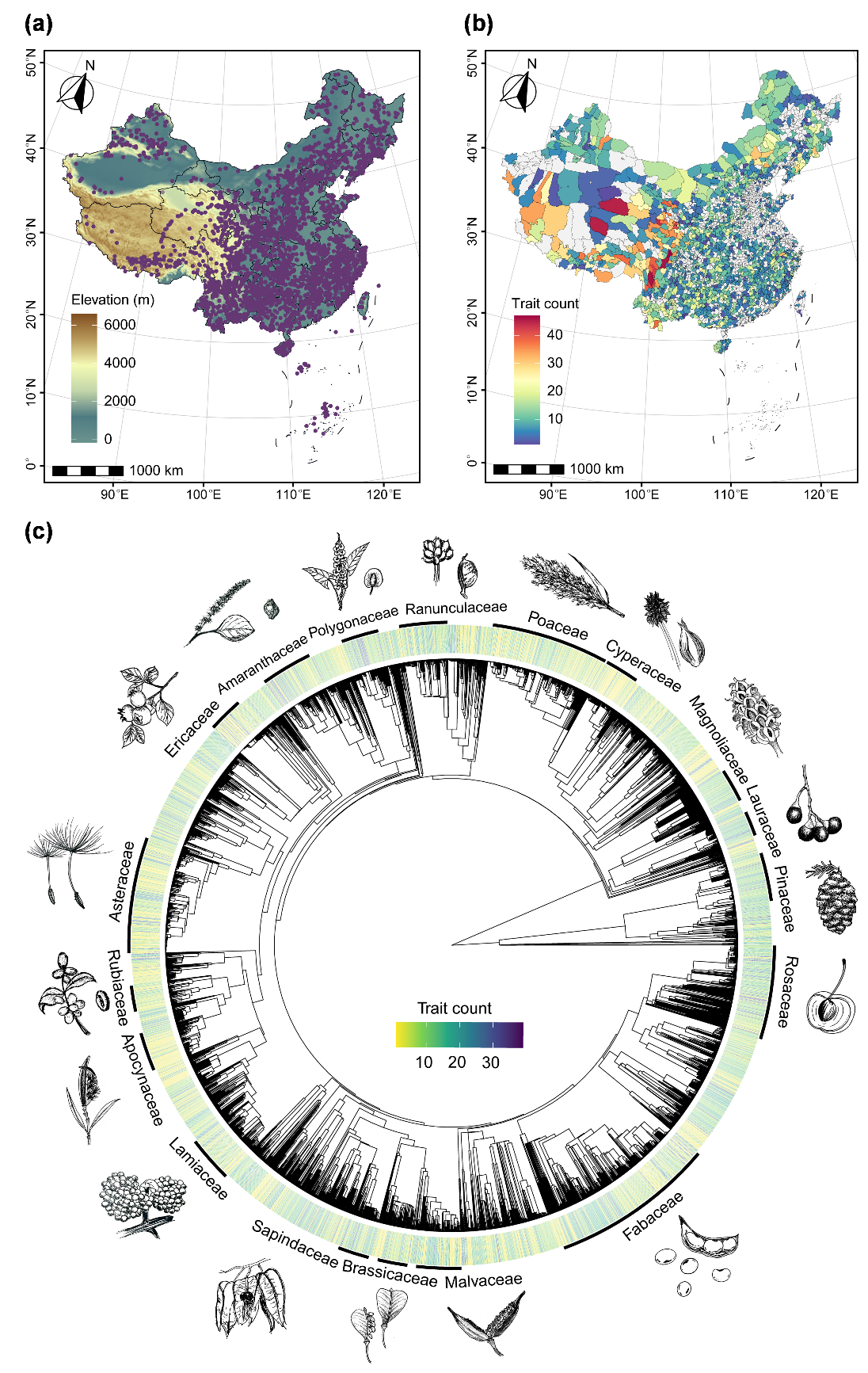 中国種子形質データベースにおける地理と系統発生にわたる形質の網羅性(画像提供:WANG Haoyu)
中国種子形質データベースにおける地理と系統発生にわたる形質の網羅性(画像提供:WANG Haoyu)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202506/t20250603_1044937.shtml
- https://nph.onlinelibrary.wiley.com/doi/10.1111/nph.70296
中国種子形質データベース:中国植物の胞子形質に関するキュレーションリソース Chinese Seed Trait Database: a curated resource for diaspore traits in the Chinese flora
Hao-Yu Wang, Xue-Lin Chen, Si-Chong Chen
New Phytologist Published: 12 June 2025
DOI:https://doi.org/10.1111/nph.70296
Current state of the Chinese Seed Trait Database (CSTD)
CSTD has so far been assembled from 694 sources in the Chinese language, including 681 journal papers, 10 books, and three online datasets. In total, CSTD comprises 110 451 records across 118 distinct seed traits for 3897 species in 1416 genera and 214 families, covering a broad range of climates and biomes with most records geo-referenced. We conducted an extensive literature search in the China National Knowledge Infrastructure and the Taiwan Airiti Library, using the keywords ‘seed’ and ‘trait’ in Chinese. The search included published literature from 1980 to 2024, ultimately resulting in a total of 8443 papers, of which 681 were identified as containing seed trait data. Meanwhile, we digitised data from 10 books, which were published by Chinese scholars based on their specific work. We also acquired relevant data from three online datasets in Chinese deposited in the Plant Science Data Centre (https://www.plantplus.cn/), Science Data Bank (http://www.scidb.cn), and Global Change Research Data Publishing & Repository (https://geodoi.ac.cn/WebEn/Default.aspx).
The database is structured as a single table, divided into four sections according to column headings (Fig. 1): (1) site information, such as sampling location, coordinates, and macroclimate; (2) species information, such as taxonomy, growth form, and life form; (3) trait information, such as trait name, trait value, value type, and unit and (4) other information, such as reference and note.

Fig. 1
Site information
We have incorporated site information whenever possible, to allow users to access both geographic and trait information on species. We retrieved detailed coordinates (WGS84) and corresponding climate data (mean annual temperature and precipitation) from the original sources whenever possible. Otherwise, for records with toponym below the county level lacking coordinates, we obtained coordinates and elevations through Google Earth or supplemented elevations using the R package elevatr (Hollister, 2023). Additionally, we extracted key climate variables from the high-precision CHELSA dataset for user convenience (Karger et al., 2017), while the provided coordinates enable users to retrieve other environmental data. Overall, 79% of the records in CSTD include toponym information, among which 66.3% have precise coordinates at county level or finer spatial resolutions (i.e. including specific information on latitude, longitude, and elevation). These geo-referenced records span a broad range across all provinces of China, with an elevational gradient from 0 to 6000 m and a latitudinal gradient from 7.4°N to 52.9°N (Fig. 2a). Notably, in addition to the extensive mainland data, 17% of the records are from islands (Fig. 2a). Counties in the Qinghai-Tibet Plateau and the Hengduan Mountains exhibit the greatest diversity of trait types, despite most data being for eastern China (Fig. 2b). The extensive geographic coverage in CSTD enables access to environmental and biotic data from additional sources, and thereby enhances efforts to investigate large-scale patterns and the underlying mechanisms.

Fig. 2
Species information
In most cases, source papers provided both Chinese and Latin names, which we standardised to accepted names using the World Checklist of Vascular Plants via the R package taxize (Chamberlain et al., 2020). For sources reporting only Chinese names, we manually converted them to Latin binomials according to the Flora of China before proceeding with standardisation. We maintained the Chinese names for species, to make CSTD a valuable resource for both international and local researchers. Plant growth forms and life forms were categorised and matched primarily based on the dataset compiled by Zheng et al. (2024). For instance, growth forms were classified based on stem lignification as herbaceous or woody, with woody plants further divided into trees and shrubs according to trunk presence and height. In total, trait coverage is relatively even across the phylogeny, with a substantial portion of species (38.7%) documented with more than 10 distinct traits, presenting comprehensive profiles (Fig. 2c). Fabaceae, Asteraceae, Poaceae, and Rosaceae have the highest species representation (Fig. 2c). However, certain species-rich families, such as Orchidaceae and Caryophyllaceae, remain underrepresented, highlighting the need for further research on their seed traits (Fig. 2c). Species in CSTD represent nearly all growth and life forms, with medium trees and deciduous species predominating among woody plants, and forbs and perennials predominating among herbaceous plants (Fig. S1). Therefore, the broad taxonomic coverage and diversity of plant forms provide a strong foundation for large-scale phylogenetic comparative analyses of seed traits, which allow us to quantify the role of seed trait syndromes in plant diversification.
Trait information
We compiled 102 continuous and 16 categorical traits, adhering to the standard definition of ‘trait’ as a stable and measurable property of an organism that reflects its response to environmental variations (Violle et al., 2007). Although morphological traits dominate the database (76% of all records), CSTD also includes a range of quantitative, phenological, physiological, chemical, and dispersal traits, which together account for c. 24% of all records, including well-documented traits, such as fruiting month, seed germination percentage, seed number per fruit, and seed dehydration tolerance (Fig. 1; Table S2). These traits, which are underrepresented in global databases such as TRY and GIFT, span all major axes of the seed trait spectrum, thereby enhancing efforts to quantify the seed functional trait spectrum (Saatkamp et al., 2019). Continuous traits comprise the majority of the CSTD, accounting for 76% of the records. We have retained the measurement units used in the data sources, so users should be aware of potential unit inconsistencies and the necessity for unit standardisation. Notably, 10 traits – seed length, seed mass, seed width, seed thickness, fruit length, fruit width, fruiting month, plant height, seed number per fruit, and seed germination percentage – account for more than half of CSTD records. These traits span 4–10 orders of magnitude; for example, seed mass exhibits the greatest variation, ranging from 0.00006 mg to 157 000 mg (Fig. S2). However, certain traits, such as seed nutrient contents (fewer than 25 records), remain severely underrepresented despite their importance, highlighting the urgent need for further research. The broad ranges of trait values and types highlight the extensive diversity of strategies captured in our database, facilitating the synthesis of primary seed trait axes across ecological scales and contributing to the comprehensive mapping of the regenerative spectrum across plant species.
Data accessibility, curation, and outlook
To enhance the visualisation of the database, we have developed the CSTD application using the shiny package (Chang et al., 2022), which is accessible at http://macroecologygroup.shinyapps.io/CSTD. Our database comprises a primary xlsx file, an R markdown script, and a visual application. The xlsx file, entitled CSTD_v1.xlsx, contains seed trait data (sheet name ‘Seed trait data’), original data sources (sheet name ‘Reference’), metadata of column names (sheet name ‘Data description’), and definitions of seed traits (sheet name ‘Trait description’). The R markdown file provides code for taxonomic standardisation and figure generation. By visiting the application, users can easily access the seed trait data as well as the original sampling location for each species. Additionally, to promote open access and facilitate data use, the CSTD files are also available in the Figshare data repository (https://doi.org/10.6084/m9.figshare.28152035). For the convenience of Chinese-speaking users, a summary of this work in Chinese is provided in Notes S1.
We are dedicated to keeping CSTD updated with the latest data, while continuously enhancing its accuracy and comprehensiveness. Specifically, we will continue curating CSTD by incorporating new Chinese publications and further expanding the literature synthesis to complement relevant data from international sources. Furthermore, we are going to expand CSTD to cover previously neglected plant lineages or seed traits through field sampling and trait measurements. We also invite the global plant science community to contribute seed trait data for the Chinese flora. Despite each record having been thoroughly checked for accuracy, some errors may still remain. We encourage users to actively contribute to the ongoing maintenance and improvement of the database by offering feedback and suggestions.
In summary, CSTD mitigates the scarcity of seed trait data by bridging valuable knowledge hidden in literature published in the Chinese language to the whole plant science community. We envision that this work represents an important step towards a global seed trait database, hopefully fostering more robust studies in the plant sciences. Our database not only allows for in-depth research on specific species but also provides a framework for comparative studies that can provide insights into large-scale trends in plant regenerative strategies and their ecological consequences. CSTD enhances the potential for cross-taxon and cross-geography analyses and meta-analyses aimed at testing ecological hypotheses related to plant regeneration from seeds, yet also goes a long way towards addressing the Raunkiæran shortfall.
Acknowledgements
S-CC was supported by the National Natural Science Foundation of China (32371612) and a start-up research grant from Wuhan Botanical Garden (E1559902). We thank undergraduate interns Xu-Ze Dong, Yi-Hao Yuan, Zi-Rui Chen, Wen-Jun He, Jia-Ze Li, and Yu-Qi Jin for their assistance in data collection, Lan Wu for botanical illustration, Tian-Hao Xia and Lin Xie for data examination, Carol Baskin and Jerry Baskin for beneficial discussion. We also thank the editor and anonymous reviewers for their constructive suggestions, which helped improve the manuscript.