2022-08-16 ペンシルベニア州立大学(PennState)
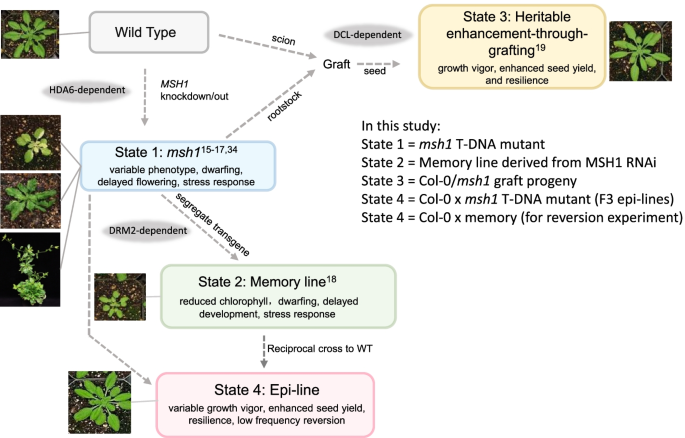
研究チームは、MSH1遺伝子を操作して、植物のストレス応答と生育力に影響を与える少なくとも4つの異なる非遺伝的状態を引き起こした。そして、これらの4つの状態から得られたデータを相互に比較することで、植物の成長に関連するデータを解読できる、ゲノム内の特定のエピジェネティック変化の標的遺伝子を特定した。
この遺伝子を操作すると、植物の外見や成長特性が変化し、野生型シロイヌナズナよりも成長が遅く、開花が遅れ、成長力が強くなり、種子の着生が増えるなど、植物の生存に有利となる可能性があると報告された。
<関連情報>
- https://www.psu.edu/news/research/story/plant-molecular-geneticists-discover-and-begin-crack-epigenetic-code/
- https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02731-w
シロイヌナズナMSH1システムにおけるRdDMを介した初期化効果のメチローム解読 Methylome decoding of RdDM-mediated reprogramming effects in the Arabidopsis MSH1 system
Hardik Kundariya,Robersy Sanchez,Xiaodong Yang,Alenka Hafner & Sally A. Mackenzie
Genome Biology Published:04 August 2022
DOI:https://doi.org/10.1186/s13059-022-02731-w
Abstract
Background
Plants undergo programmed chromatin changes in response to environment, influencing heritable phenotypic plasticity. The RNA-directed DNA methylation (RdDM) pathway is an essential component of this reprogramming process. The relationship of epigenomic changes to gene networks on a genome-wide basis has been elusive, particularly for intragenic DNA methylation repatterning.
Results
Epigenomic reprogramming is tractable to detailed study and cross-species modeling in the MSH1 system, where perturbation of the plant-specific gene MSH1 triggers at least four distinct nongenetic states to impact plant stress response and growth vigor. Within this system, we have defined RdDM target loci toward decoding phenotype-relevant methylome data. We analyze intragenic methylome repatterning associated with phenotype transitions, identifying state-specific cytosine methylation changes in pivotal growth-versus-stress, chromatin remodeling, and RNA spliceosome gene networks that encompass 871 genes. Over 77% of these genes, and 81% of their central network hubs, are functionally confirmed as RdDM targets based on analysis of mutant datasets and sRNA cluster associations. These dcl2/dcl3/dcl4-sensitive gene methylation sites, many present as singular cytosines, reside within identifiable sequence motifs. These data reflect intragenic methylation repatterning that is targeted and amenable to prediction.
Conclusions
A prevailing assumption that biologically relevant DNA methylation variation occurs predominantly in density-defined differentially methylated regions overlooks behavioral features of intragenic, single-site cytosine methylation variation. RdDM-dependent methylation changes within identifiable sequence motifs reveal gene hubs within networks discriminating stress response and growth vigor epigenetic phenotypes. This study uncovers components of a methylome “code” for de novo intragenic methylation repatterning during plant phenotype transitions.


