2024-05-13 シンガポール国立大学(NUS)
<関連情報>
- https://news.nus.edu.sg/macrophage-variations-linked-to-patients-survival-in-cancer/
- https://www.nature.com/articles/s41467-024-46220-z
びまん性大細胞型B細胞リンパ腫におけるマクロファージの不均一性と予後の重要性を明らかにする空間分解トランスクリプトミクス Spatially-resolved transcriptomics reveal macrophage heterogeneity and prognostic significance in diffuse large B-cell lymphoma
Min Liu,Giorgio Bertolazzi,Shruti Sridhar,Rui Xue Lee,Patrick Jaynes,Kevin Mulder,Nicholas Syn,Michal Marek Hoppe,Fan Shuangyi,Yanfen Peng,Jocelyn Thng,Reiya Chua,Jayalakshmi,Yogeshini Batumalai,Sanjay De Mel,Limei Poon,Esther Hian Li Chan,Joanne Lee,Susan Swee-Shan Hue,Sheng-Tsung Chang,Shih-Sung Chuang,K. George Chandy,Xiaofei Ye,Qiang Pan-Hammarström,… Anand D. Jeyasekharan
Nature Communications Published:08 March 2024
DOI:https://doi.org/10.1038/s41467-024-46220-z
Abstract
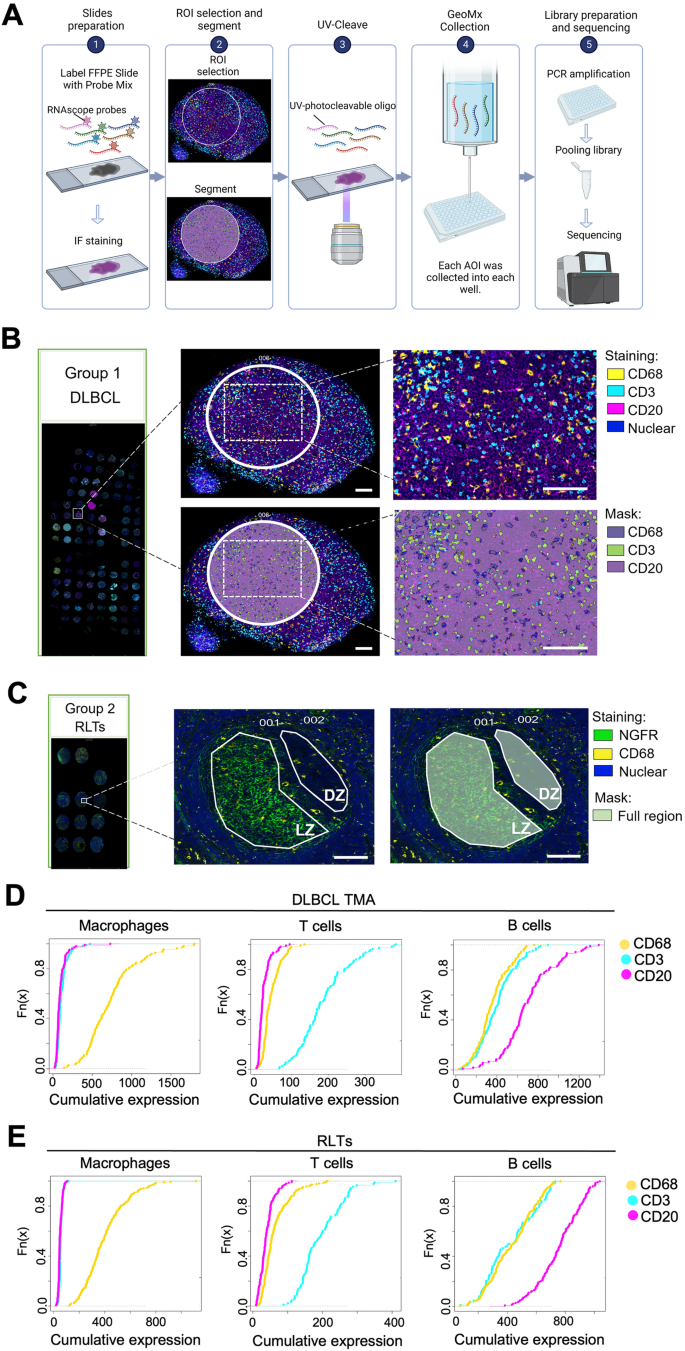
Macrophages are abundant immune cells in the microenvironment of diffuse large B-cell lymphoma (DLBCL). Macrophage estimation by immunohistochemistry shows varying prognostic significance across studies in DLBCL, and does not provide a comprehensive analysis of macrophage subtypes. Here, using digital spatial profiling with whole transcriptome analysis of CD68+ cells, we characterize macrophages in distinct spatial niches of reactive lymphoid tissues (RLTs) and DLBCL. We reveal transcriptomic differences between macrophages within RLTs (light zone /dark zone, germinal center/ interfollicular), and between disease states (RLTs/ DLBCL), which we then use to generate six spatially-derived macrophage signatures (MacroSigs). We proceed to interrogate these MacroSigs in macrophage and DLBCL single-cell RNA-sequencing datasets, and in gene-expression data from multiple DLBCL cohorts. We show that specific MacroSigs are associated with cell-of-origin subtypes and overall survival in DLBCL. This study provides a spatially-resolved whole-transcriptome atlas of macrophages in reactive and malignant lymphoid tissues, showing biological and clinical significance.