2024-10-31 ミュンヘン大学(LMU)
乳がんの転移に関する包括的研究で、転移細胞の多様性とそれらが周囲細胞とどのように相互作用するかが解析されました。この研究はRNA解析と空間的発現プロファイリングを組み合わせ、多様な手法で異なる転移細胞の特徴を明らかにしました。研究により、異なる患者間での発現プロファイルの違いや、特定の免疫細胞が腫瘍に近い場合の遺伝子発現の変化が確認され、将来的に転移乳がん治療の改善に寄与する可能性が示されました。
<関連情報>
- https://www.lmu.de/en/newsroom/news-overview/news/breast-cancer-the-diversity-of-metastases.html
- https://www.nature.com/articles/s41591-024-03215-z
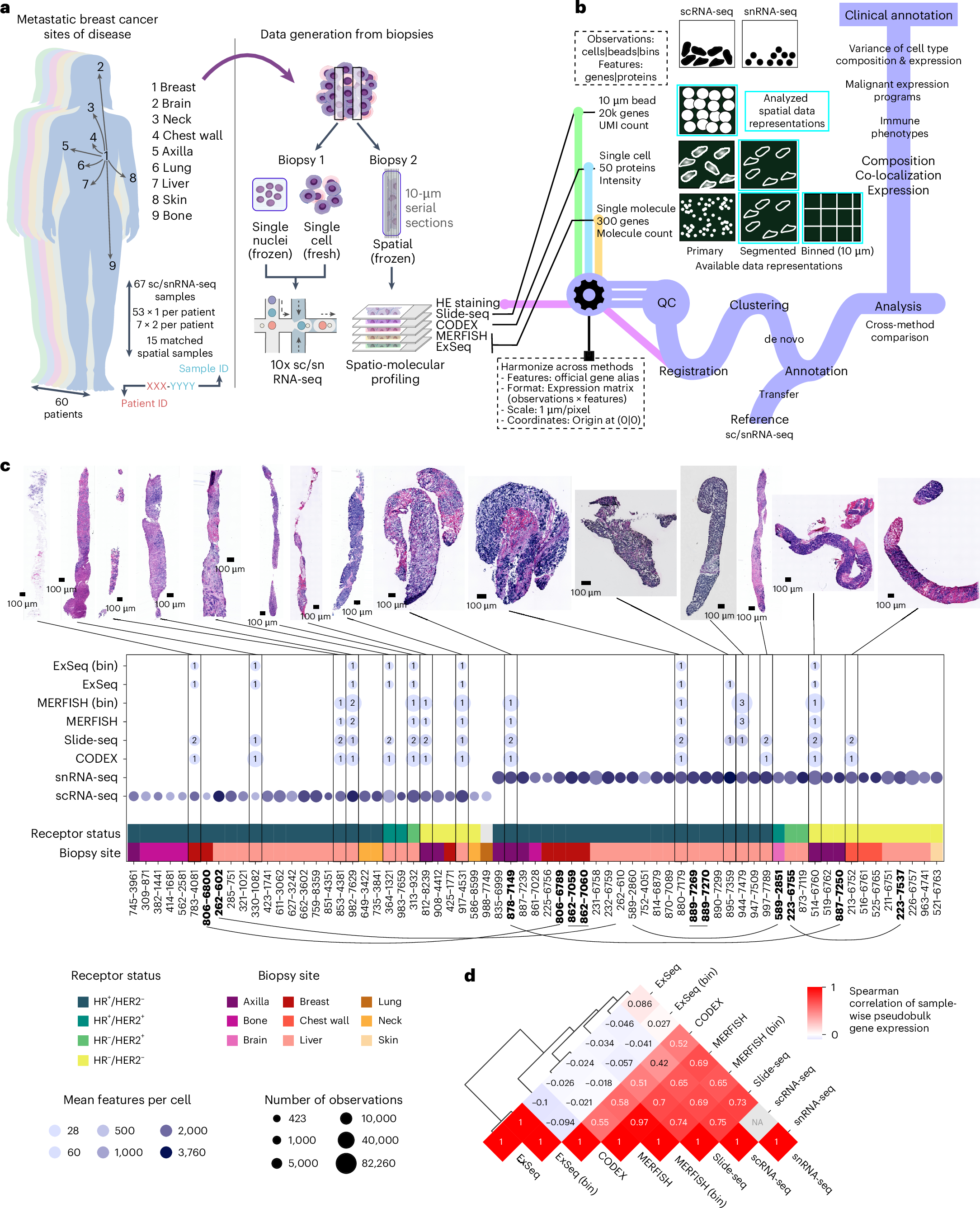
臨床病理学的特徴を横断する転移性乳癌生検のマルチモーダル単一細胞・空間発現マップ A multi-modal single-cell and spatial expression map of metastatic breast cancer biopsies across clinicopathological features
Johanna Klughammer,Daniel L. Abravanel,Åsa Segerstolpe,Timothy R. Blosser,Yury Goltsev,Yi Cui,Daniel R. Goodwin,Anubhav Sinha,Orr Ashenberg,Michal Slyper,Sébastien Vigneau,Judit Jané‐Valbuena,Shahar Alon,Chiara Caraccio,Judy Chen,Ofir Cohen,Nicole Cullen,Laura K. DelloStritto,Danielle Dionne,Janet Files,Allison Frangieh,Karla Helvie,Melissa E. Hughes,Stephanie Inga,… Nikhil Wagle
Nature Medicine Published:30 October 2024
DOI:https://doi.org/10.1038/s41591-024-03215-z
Abstract
Although metastatic disease is the leading cause of cancer-related deaths, its tumor microenvironment remains poorly characterized due to technical and biospecimen limitations. In this study, we assembled a multi-modal spatial and cellular map of 67 tumor biopsies from 60 patients with metastatic breast cancer across diverse clinicopathological features and nine anatomic sites with detailed clinical annotations. We combined single-cell or single-nucleus RNA sequencing for all biopsies with a panel of four spatial expression assays (Slide-seq, MERFISH, ExSeq and CODEX) and H&E staining of consecutive serial sections from up to 15 of these biopsies. We leveraged the coupled measurements to provide reference points for the utility and integration of different experimental techniques and used them to assess variability in cell type composition and expression as well as emerging spatial expression characteristics across clinicopathological and methodological diversity. Finally, we assessed spatial expression and co-localization features of macrophage populations, characterized three distinct spatial phenotypes of epithelial-to-mesenchymal transition and identified expression programs associated with local T cell infiltration versus exclusion, showcasing the potential of clinically relevant discovery in such maps.


