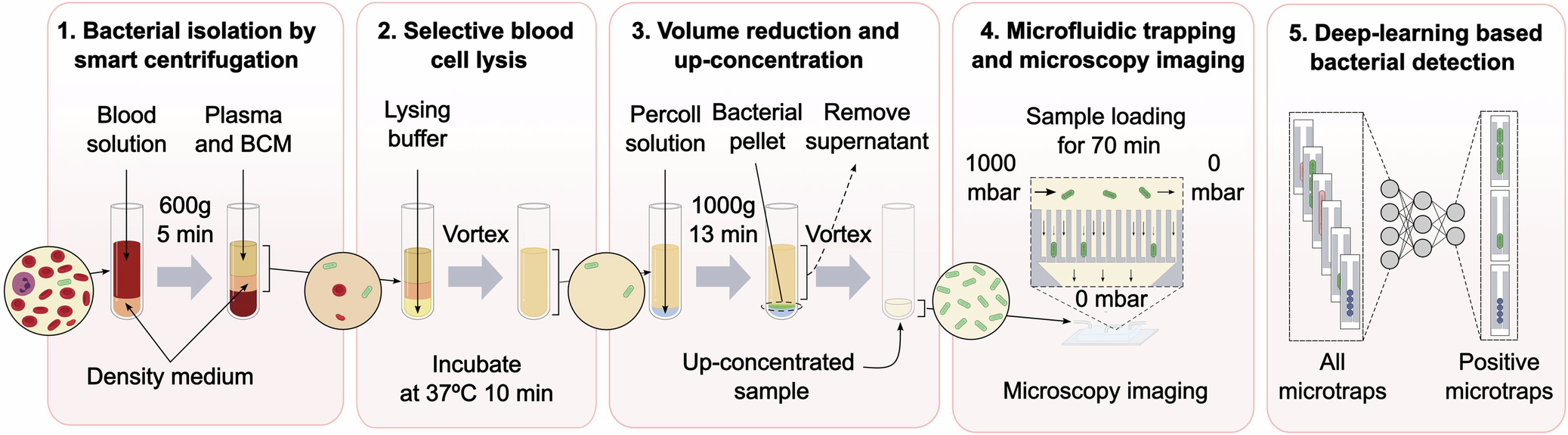
2025-08-28 ゲーテ大学
Web要約 の発言:
<関連情報>
- https://aktuelles.uni-frankfurt.de/english/how-plants-rot-new-method-decodes-hidden-decomposers-of-wood-and-leaves/
- https://academic.oup.com/mbe/article/42/6/msaf120/8158454
fDOGを用いた特徴構造認識型オルソログ検索が明らかにする、植物細胞壁分解酵素の生命圏における分布 Feature Architecture-Aware Ortholog Search With fDOG Reveals the Distribution of Plant Cell Wall-Degrading Enzymes Across Life
Vinh Tran , Felix Langschied , Hannah Muelbaier , Julian Dosch , Freya Arthen , Miklos Balint , Ingo Ebersberger
Molecular Biology and Evolution Published:09 June 2025
DOI:https://doi.org/10.1093/molbev/msaf120
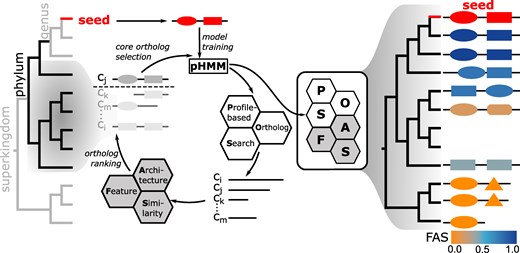
Abstract
The decomposition of plant material is a key driver of the global carbon cycle, traditionally attributed to fungi and bacteria. However, some invertebrates also possess orthologs to bacterial or fungal cellulolytic enzymes, likely acquired via horizontal gene transfer. This reticulated mode of evolution necessitates ortholog searches in large taxon sets to comprehensively map the repertoire of plant cell wall-degrading enzymes (PCDs) across the tree of life, a task surpassing capacities of current software. Here, we use fDOG, a novel profile-based ortholog search tool to trace 235 potential PCDs across more than 18,000 taxa. fDOG allows to start the ortholog search from a single protein sequence as a seed, it performs on par with state-of-the-art software that require the comparison of entire proteomes, and it is unique in routinely scoring protein feature architecture differences between the seed protein and its orthologs. Visualizing the presence–absence patterns of PCD orthologs using a Uniform Manifold Approximation and Projection highlights taxa where recent changes in the enzyme repertoire indicate a change in lifestyle. Three invertebrates have a particularly rich set of PCD orthologs encoded in their genome. Only few of the orthologs show differing protein feature architectures relative to the seed that suggest functional modifications. Thus, the corresponding species represent lineages within the invertebrates that may contribute to the global carbon cycle. This study shows how fDOG can be used to create a multi-scale view on the taxonomic distribution of a metabolic capacity that ranges from tree of life-wide surveys to individual feature architecture changes within a species.