2025-09-12 ロックフェラー大学
<関連情報>
- https://www.rockefeller.edu/news/38239-hundreds-of-new-bacteria-and-two-potential-antibiotics-found-in-soil/
- https://www.nature.com/articles/s41587-025-02810-w
土壌メタゲノムのテラベース規模ロングリードシーケンシングにより発見された生物活性分子 Bioactive molecules unearthed by terabase-scale long-read sequencing of a soil metagenome
Ján Burian,Robert E. Boer,Yozen Hernandez,Adrian Morales-Amador,Linhai Jiang,Abir Bhattacharjee,Cecilia Panfil,Melinda A. Ternei & Sean F. Brady
Nature Biotechnology Published:12 September 2025
DOI:https://doi.org/10.1038/s41587-025-02810-w
Abstract
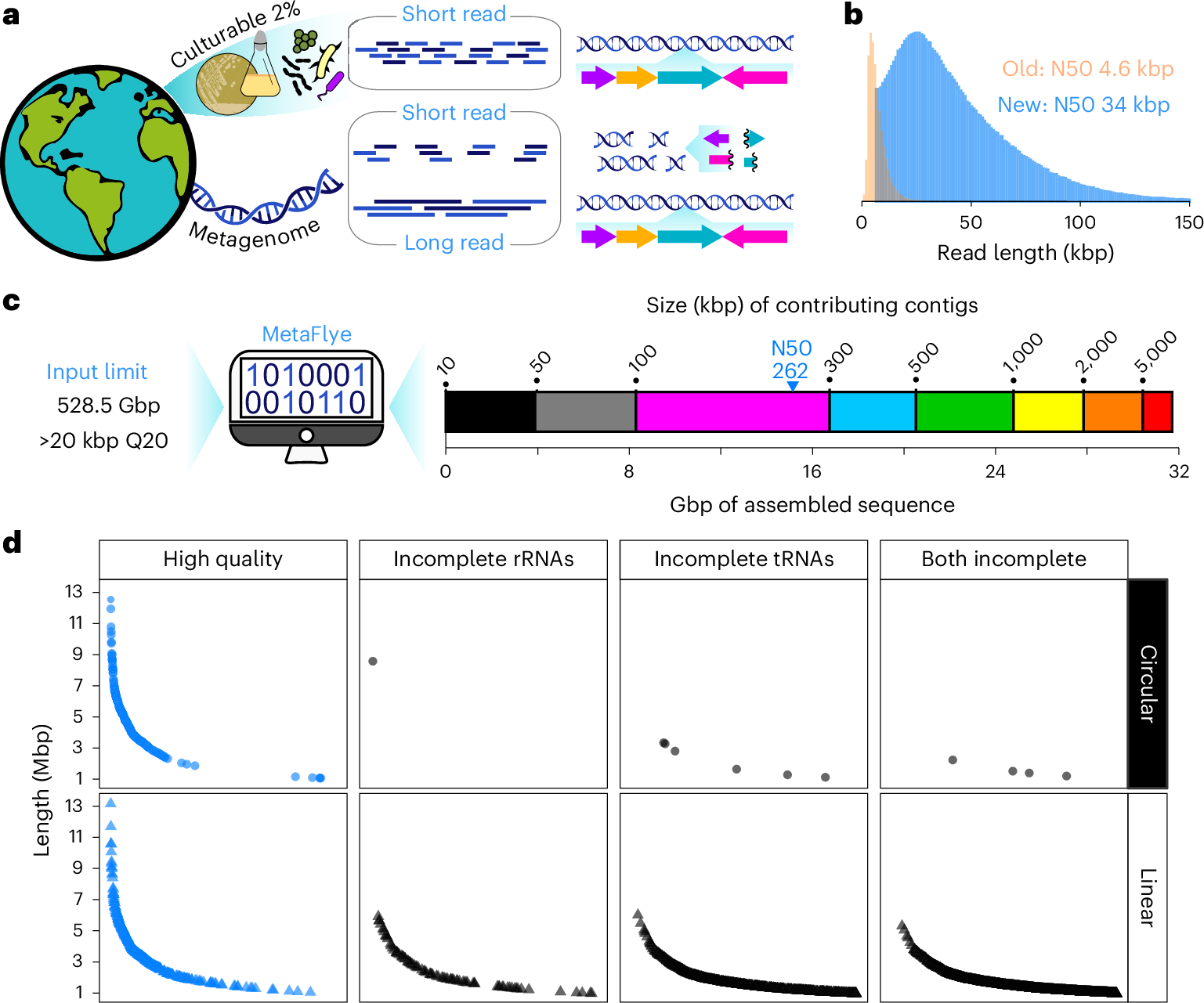
Metagenomics provides access to the genetic diversity of uncultured bacteria through analysis of DNA extracted from whole microbial communities. Long-read sequencing is advancing metagenomic discovery by generating larger DNA assemblies than previously possible. However, harnessing the potential of long-read sequencing to access the vast diversity within soil microbiomes is hampered by the challenge of isolating high-quality DNA. Here we introduce a method that can liberate large, high-quality metagenomic DNA fragments from soil bacteria and pair them with optimized nanopore long-read sequencing to generate megabase-sized assemblies. Using this method, we uncover hundreds of complete circular metagenomic genomes from a single soil sample. Through a combination of bioinformatic prediction and chemical synthesis, we convert nonribosomal peptide biosynthetic gene clusters directly into bioactive molecules, identifying antibiotics with rare modes of action and activity against multidrug-resistant pathogens. Our approach advances metagenomic access to the vast genetic diversity of the uncultured bacterial majority and provides a means to convert it to bioactive molecules.


