2025-09-24 パシフィック・ノースウェスト国立研究所(PNNL)
<関連情報>
- https://www.pnnl.gov/publications/redox-driven-protein-complexes-signal-metabolic-modulation-cyanobacteria
- https://journals.aps.org/prxlife/abstract/10.1103/l2dp-kw2t
マルチオミクス解析により、光撹乱下におけるSynechococcus Elongatus PCC 7942 の炭素代謝の時間スケールが明らかになった Multi-Omics Reveals Temporal Scales of Carbon Metabolism in Synechococcus Elongatus PCC 7942 Under Light Disturbance
Connah G. M. Johnson, Zachary Johnson, Liam S. Mackey, Xiaolu Li, Natalie C. Sadler, Tong Zhang, Wei-Jun Qian, Pavlo Bohutskyi, Song Feng, et al.
PRX Life Published: 2 September, 2025
DOI: https://doi.org/10.1103/l2dp-kw2t
Abstract
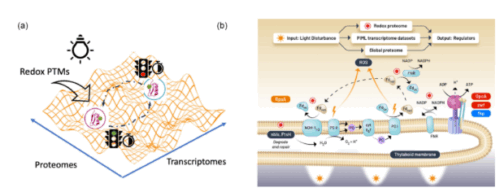
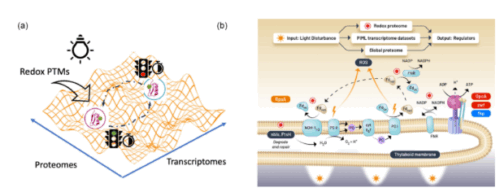
Central carbon metabolism in model cyanobacteria involves multiple pathways to adapt to energy-light limitations across diel cycles. However, the success in mechanistic modeling for phenotypic prediction of the protein regulators in the metabolic state depends on capturing the vast possibilities emerging from multiple regulatory pathways in complex biological processes. Here, we developed a physics-informed machine learning approach based on energy-landscape concepts to predict regulatory proteins responding to cyclic circadian and unforeseen light perturbations in cyanobacterial metabolic networks. Our approach provides interpretable de novo models for inferring gene expression dynamics from Synechococcus elongatus over diel cycles and using redox proteome analysis to distinguish immediate light-responsive elements from circadian-regulated processes in carbon metabolism pathways. We identified distinct temporal signatures with the analysis of the redox proteome: there was an immediate shift in cysteine redox states accompanied by a limited change in protein abundance under constant illumination and after 2 hours of darkness. This discovery indicates that the generation of reductants coordinates photoinduced electron transport with redox metabolic pathways in two discernable molecular mechanisms: fast redox-based protein modifications occur immediately after the light disturbance, followed by slow transcriptional regulations across networks. This temporal regulation reveals how metabolic networks integrate rapid light responses with programmed circadian rhythms to maintain cellular homeostasis under the light-energy limitations over the diel cycle.