2025-10-30 カリフォルニア工科大学(Caltech)
Web要約 の発言:
![]()
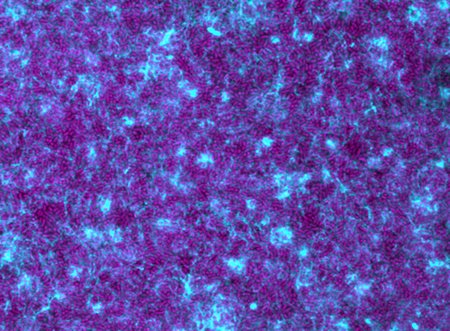
Bacteria, labeled in magenta, form a massive biofilm community connected by a gooey extracellular matrix (blue).Credit: G. Squyres
<関連情報>
- https://www.caltech.edu/about/news/a-new-perspective-on-bacterial-biofilm-defenses
- https://www.pnas.org/doi/10.1073/pnas.2514210122
緑膿菌バイオフィルムのマトリックスにおけるeDNAの形態形成は単細胞溶解パターンに依存する Single-cell lysis patterns morphogenesis of eDNA in the matrix of Pseudomonas aeruginosa biofilms
Georgia R. Squyres and Dianne K. Newman
Proceedings of the National Academy of Sciences Published:October 6, 2025
DOI:https://doi.org/10.1073/pnas.2514210122
Significance
Most bacteria on earth live in groups called biofilms. Biofilms behave differently than their constituent bacteria, including exhibiting increased antimicrobial tolerance. Within biofilms, bacteria spatiotemporally pattern their behavior, but how this patterning is organized and how it leads to these emergent biofilm-scale properties is unclear. Using extracellular DNA (eDNA) release as a model process for understanding the interplay between individual behavior and collective function in 4 dimensions, here we consider what governs eDNA release in Pseudomonas aeruginosa biofilms and its consequences for biofilm development. We find that cell lysis is patterned by individual biofilm cells following simple rules about where and when to lyse, and that elucidating these rules can explain how biofilm-scale properties like matrix morphogenesis arise from single-cell behaviors.
Abstract
When bacteria form a biofilm, complex behaviors emerge. Biofilm bacteria differ from their free-living counterparts, exhibiting heterogenous, spatiotemporally patterned behavior. Can we explain these patterns by defining the rules that govern single-cell behavior in biofilms? By understanding these rules, can we explain emergent functions at the biofilm scale? Here we reveal how the architecture of extracellular DNA (eDNA) in the biofilm matrix is controlled by single-cell lysis during Pseudomonas aeruginosa biofilm development. We extend single-cell imaging methods to capture complete biofilm development over 5+ d, characterizing the stages of biofilm development and visualizing eDNA matrix morphogenesis from start to finish. Mapping the spatiotemporal distribution of single-cell lysis events shows that cell lysis is spatiotemporally patterned, concentrated in a region 5 µm below the biofilm surface that moves with the biofilm as it grows. Using analytical modeling, we examined the consequences of patterning at the biofilm scale. Cell lysis patterning defines eDNA in the matrix: Patterned lysis is sufficient to explain the final eDNA distribution. Cell lysis and biofilm growth are coupled such that the amount of eDNA in the biofilm scales with its volume; this patterning results in a predominantly uniform eDNA matrix architecture, which could not occur without patterning. Finally, we find that patterning of cell lysis is self-organized by nutrient gradients, with maximal lysis occurring in regions where oxygen is present and carbon is limited. The ability of cells to use self-generated nutrient gradients as positioning cues to establish depth-based patterning is a striking feature of bacterial biofilm development.