2022-10-14 アルゴンヌ国立研究所(ANL)
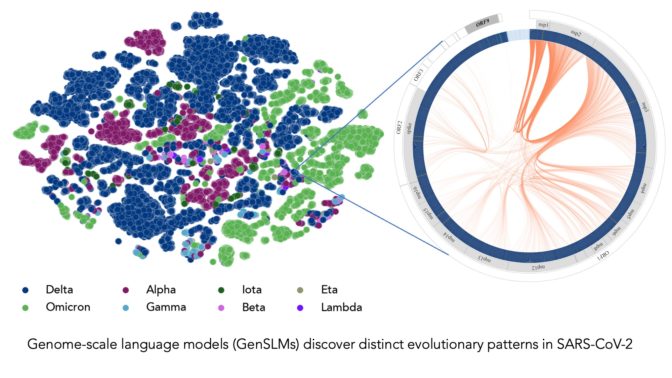
Trained on a year’s worth of SARS-CoV-2 genome data, the model can infer the distinction between various viral strains. Each dot on the left corresponds to a sequenced SARS-CoV-2 viral strain, color-coded by variant. The figure on the right zooms into one particular strain of the virus, which captures evolutionary couplings across the viral proteins specific to this strain. Image courtesy of Argonne National Laboratory’s Bharat Kale, Max Zvyagin and Michael E. Papka.
共同研究者たちは、LLM が約 1,500 個のヌクレオチドの長い文字列を文章のように扱えるように、階層的拡散手法を設計した。
研究チームはまず、Bacterial and Viral Bioinformatics Resource Centerのオープンソースデータを使用して、細菌のような単細胞生物である原核生物の1億1000万以上の遺伝子配列でLLMを事前学習させた。その後、COVIDウイルスの高品質なゲノム配列150万件を用いて、モデルの微調整を行った。
さらに、より広範なデータセットで事前学習を行うことで、将来のプロジェクトで他の予測タスクにも対応できるよう、モデルの汎化を図った。
COVIDのデータで微調整を行った結果、LLMはウイルスの変異体のゲノム配列を識別することができた。また、独自の塩基配列を生成することもでき、COVIDゲノムの潜在的な変異を予測することで、将来懸念される変異を科学者が予測するのに役立てることができた。
<関連情報>
- https://blogs.nvidia.com/blog/2022/11/14/genomic-large-language-model-predicts-covid-variants/
- https://www.biorxiv.org/content/10.1101/2022.10.10.511571v1
GenSLMs :ゲノムスケール言語モデルによるSARS-CoV-2の進化ダイナミクスの解明 GenSLMs: Genome-scale language models reveal SARS-CoV-2 evolutionary dynamics
Maxim Zvyagin, Alexander Brace, Kyle Hippe, Yuntian Deng, Bin Zhang, Cindy Orozco Bohorquez, Austin Clyde, Bharat Kale, Danilo Perez-Rivera, Heng Ma, Carla M. Mann, Michael Irvin, J. Gregory Pauloski, Logan Ward, Valerie Hayot, Murali Emani, Sam Foreman, Zhen Xie, Diangen Lin, Maulik Shukla, Weili Nie, Josh Romero, Christian Dallago, Arash Vahdat, Chaowei Xiao, Thomas Gibbs, Ian Foster, James J. Davis, Michael E. Papka, Thomas Brettin, Rick Stevens, Anima Anandkumar, Venkatram Vishwanath, Arvind Ramanathan
bio Rxiv Posted:October 11, 2022.
DOI:https://doi.org/10.1101/2022.10.10.511571
ABSTRACT
Our work seeks to transform how new and emergent variants of pandemic causing viruses, specially SARS-CoV-2, are identified and classified. By adapting large language models (LLMs) for genomic data, we build genome-scale language models (GenSLMs) which can learn the evolutionary landscape of SARS-CoV-2 genomes. By pretraining on over 110 million prokaryotic gene sequences, and then finetuning a SARS-CoV-2 specific model on 1.5 million genomes, we show that GenSLM can accurately and rapidly identify variants of concern. Thus, to our knowledge, GenSLM represents one of the first whole genome scale foundation models which can generalize to other prediction tasks. We demonstrate the scaling of GenSLMs on both GPU-based supercomputers and AI-hardware accelerators, achieving over 1.54 zettaflops in training runs. We present initial scientific insights gleaned from examining GenSLMs in tracking the evolutionary dynamics of SARS-CoV-2, noting that its full potential on large biological data is yet to be realized.